Abstract
Ottelia cordata is a typical submerged plant to the family of hydrocharitaceae. In this work, complete chloroplast genome of O. cordata was assembled and characterized from high-throughput Illumina sequencing data. The chloroplasgenome of O. cordatais 157,869bp in length. The O. cordata cp genome encodes 128 functional genes, including 85 protein-coding genes, 35 tRNA genes, and 8 rRNA genes. The GC content of the genome was 36,57%. Its nucleotide composition is asymmetric with an overall A + T content of 63.44%. Phylogenetic analysis indicated that O. cordata is closely related to the Elodea. These results could offer molecular markers and helpful in understanding about the role of O. cordata evolution and protection.
Introduction
Ottelia cordata is an annual aquatic herbaceous plant, which belong to the genius Ottelia Pers of the family Hydrocharitaceae. It is only distributed in Hainan province in China and also in Myanma, Thailand, and Cambodia (Xu et al. Citation2010). Ottelia cordatawas endangered in recent years due to various environmental threats, such as wetland destruction, invasion of alien species and water pollution. In this work, we assembled and characterized O. cordata complete cp genome. These results were beneficial for resources study and protection of O. cordata.
The fresh O. cordata leaves were collected from Zuntan, Haikou (110°17′1.97″E, 19°47′14.43″N). The specimen was deposited at Hainan Academy of Ocean and Fisheries Sciences (P-2019-ZTO1). Total DNA was extracted using Ezup DNA kit and sequenced with Illumina Hiseq 4000. Ottelia cordata cp genome sequence was assembled by SPAdes v.3.5.0 and annotated by DOGMA (Wyman et al. Citation2004). The tRNA was annotated by the method of tRNAscan-SE2.0 (Lowe and Chan Citation2016) and ARWEN (Laslett and Canback Citation2008). The size of the O. cordata cp genome was 157,869 bp (Genbank accession number: MN056354), which has 10 bp difference with 157,879 bp in Wang’s research (Wang et al., Citation2019). This genome included a LSC region of 87,672 bp, a SSC region of 19,093 bp, and a pair of IRs (IRa and IRb) of 25,552bp each. Totally, 128 functional genes were identified in O. cordata, including 85 protein-coding genes, 35 tRNA genes, and 8 rRNA genes. Totally, 44 SSRs and 43 pairs of large repeat sequences were found in this genome. Among the 44 SSRs, 14 SSRs were located in protein-coding genes (atpF, rpoC2, rpoC1, accD, clpP, rps8 and ycf1). Among these, 43 pairs of long repeat sequences, 18 long repeat sequences were located in protein-coding genes (psaA, ycf2).
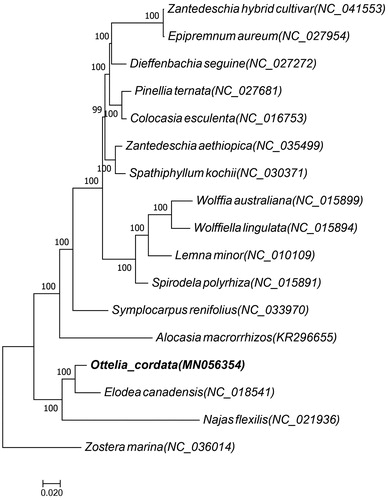
In order to ascertain its phylogenetic placement location of O. cordata, 17 species respectively belong to three family of Araceae, Hydrocharitaceae, and Zosteraceae were selected and their common coding genes were analyzed with maximum likelihood (ML) methods by software RAxML8.1.5 with the model of GTR + I+G (Stamatakis, Citation2014). Among them, 13 species belong to the family of Araceae and one species belong to the family of Zosteraceae. The result supported the position of O. cordata as a sister to Elodea within the Alismatales order (). We believe that it will provide new insight into its evolutionary research and conservation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in figshare at http://doi.org/10.6084/m9.figshare.12089679.
Additional information
Funding
References
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang H-X, Guo J-L, Li Z-M, Yu Y-H, Zhang Y-H. 2019. Characterization of the complete chloroplast genome of an endangered aquatic macrophyte, Ottelia cordata (Hydrocharitaceae). Mitochondrial DNA Part B. 4(1):1839–1840.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Xu NN, Yu S, Zhang JG, Tsang PK, Chen XY. 2010. Microsatellite primers for Halophila ovalis and cross-amplification in H. minor (Hydrocharitaceae). Am J Bot. 97(6):e56–e57.