Abstract
Mangrove tree Avicenna marina has great ecological significance in maintaining coastal ecosystem, but its unique potential for gene functions and genetic diversity underlying ecological adaptation remains investigation. In this study, the chloroplast genome of A. marina was first characterized by sequencing chloroplast DNA with Illumina technology. The A. marina genome was 147,909 bp in size with a typical quadripartite structure, which was deposited in GenBank under the accession number MT108381.
Introduction
Avicennia marina, a pioneer mangrove species with the strong wind control and the higher ability of salt tolerance in China (Liao and Zhang Citation2014). It plays a significant role in supporting coastal ecosystem and holds unique potential for studying molecular mechanisms underlying ecological adaptation.
The chloroplast genomes contain highly conserved essential genes for plant growth and development (Wicke et al. Citation2011), but they also harbor a variety of genetic polymorphisms (Daniell et al. Citation2016; Sakaguchi et al. Citation2017). The whole chloroplast genome data obtained from next-generation sequencing technology offer promise for further researches in chloroplast gene functions and genome diversity.
The fresh leaves of A. marina were collected in August 2018 from Zhangjiang Estuary, a Mangrove National Nature Reserve (23°55′N, 117°26′E), Fujian, China. Voucher specimen was kept in the Herbarium of School of Life Sciences, Xiamen University with specimen code XYZ20180818. The intact chloroplasts were isolated with a high salt isolation buffer followed by a saline Percoll gradient centrifugation, from which the chloroplast DNA (cpDNA) was isolated according to Vieira et al. (Citation2014). Illumina sequencing produced 7,672,820 paired-end raw reads with an Illumina HiSeq TM2000 platform. Filtered 6,445,660 chloroplast genome reads (949,811,469 bp) were used to constructure the chloroplast genome by the NOVOPlasty program (version 3.3) with Aloysia citrodora (GenBank: NC_034695) as a reference (Dierckxsens et al. Citation2017). The web-based program GeSeq Annotator (https://chlorobox.mpimp-golm.mpg.de/geseq.html) was used to annotate protein coding genes and ribosomal RNA genes. The transfer RNA genes were identified with GeSeq and the tRNAscan-SE program v2.0.3 (Schattner et al. Citation2005). Circular representation of the chloroplast genome was created by the web-based program OGDRAW tool (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013).
The complete chloroplast genome of A. marina with a typical quadripartite structure was 147,909 bp in size (38.15% CG content), which consisted of a small single copy region (SSC) of 17,772 bp, a large single copy region (LSC) of 88,331 bp, and a pair of inverted repeats (IRs) of 20,903 bp in size. There were 112 genes in total, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes, of which 14 genes occurred in IRs, containing 5 protein-coding genes, 5 tRNA genesand 4 rRNA genes. In addition, there were 10 intron-containing genes, nine protein-coding genes and one tRNA genes.
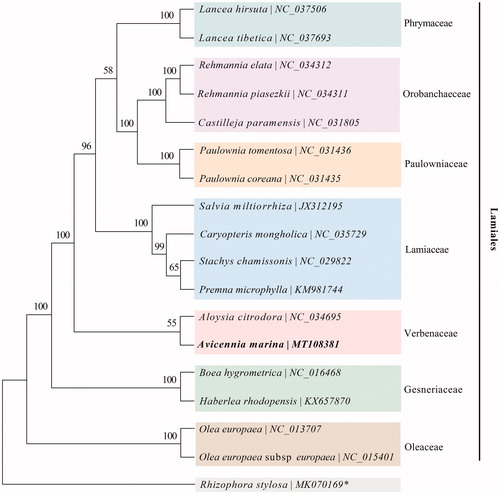
To confirm the phylogenetic position of this newly described chloroplast genome, the 16 chloroplast genomes under the order Lamiales and one mangrove species Rhizophora stylosa (as an outgroup) from NCBI were used to build the phylogenetic tree based on 76 protein-coding genes, which were aligned with MAFFT and concatenated with Gblocks (Nakamura et al., Citation2018). Phylogenetic reconstruction was based on Maximum Likelihood analysis with GTR + G nucleotide substitution Model and 1000 bootstrap replicates using the MEGA X (Kumar et al. Citation2018). The phylogenetic analysis confirmed that A. marina was grouped into the family of Verbenaceae ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank database at https://www.ncbi.nlm.nih.gov/nuccore/MT108381.
Additional information
Funding
References
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liao BW, Zhang QM. 2014. Area, distribution and species composition of mangroves in China. Wetland Sci. 12:435–440.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34(14):2490–2492.
- Sakaguchi S, Ueno S, Tsumura Y, Setoguchi H, Ito M, Hattori C, Nozoe S, Takahashi D, Nakamasu R, Sakagami T, et al. 2017. Application of a simplified method of chloroplast enrichment to small amounts of tissue for chloroplast genome sequencing. Appl Plant Sci. 5(5):1700002.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689.
- Vieira LD, Faoro H, Fraga HPD, Rogalski M, de Souza EM, Pedrosa FD, Nodari RO, Guerra MP. 2014. An improved protocol for intact chloroplasts and cpDNA isolation in Conifers. PLoS One. 9(1):e84792.
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3–5):273–297.