Abstract
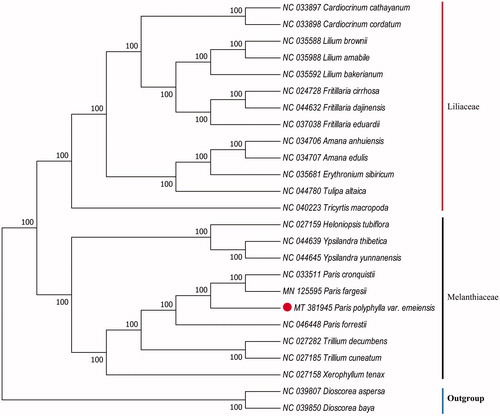
Paris polyphylla var. emeiensis H. X. Yin, H. Zhang & D. Xue is a member of the genus Paris endemic to southwest of China. The complete chloroplast (cp) genome of P. polyphylla var. emeiensis was 164,854 bp in length with 36.96% overall GC content, including a large single-copy (LSC) region of 84,438 bp and a small single-copy (SSC) region of 12,892 bp, which were separated by a pair of inverted repeats (IRs) of 33,762 bp. There were 135 genes in total, including 89 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Phylogenetic analysis indicated P. polyphylla var. emeiensis was closely related to P. fargesii and P. cronquistii.
Paris polyphylla var. emeiensis, an endemic to southwest of China, is a species of flowering herb of the family Melanthiaceae. This species was described from Mount Emei, Sichuan Province, China in 2007 for the first time (Yin et al. Citation2007). The root of P. polyphylla var. emeiensis was used as the substitute of the traditional Chinese medicine ‘Chonglou’, which have been used for the treatment of hemostasis, sore throat, parotitis, furuncle, carbuncle, snake bite, and convulsion (Fu et al. Citation2012; Liu et al. Citation2017; Duan et al. Citation2018). However, little genetic information is known about this species until now. Herein, we assembled and characterized the complete chloroplast (cp) genome of P. polyphylla var. emeiensis for the first time, and its phylogenetic analysis was investigated. The study will provide useful information for the conservation and utilization of this species.
Fresh and clean leaves of P. polyphylla var. emeiensis were sampled from Gucheng District, Lijiang, Yunnan, China (26°74′71″N, 100°27′14″E). Meanwhile, a voucher specimen was collected and deposited at the Herbarium of Dali University (20191003B2). The total DNA was extracted using the DNeasy plant mini kit (QIAGEN), and sequenced by Illumina NovaSeq system (Illumina, San Diego, CA, USA). About 7.30 Gb of raw data (51, 901, 266 reads) were assembled by NOVOPlasty (Park et al. Citation2019; Liu et al. Citation2020), and the assembled cp genome was annotated by GeSeq with default sets (Tillich et al. Citation2017; Qian et al. Citation2020). The annotated cp genome was submitted to the GenBank with the accession number of MT381945.
The cp genome of P. polyphylla var. emeiensis was 164,854 bp in length with a typical quadripartite structure of angiosperms, containing a large single-copy (LSC) region of 84,438 bp, a small single-copy (SSC) region of 12,892 bp, and a pair of inverted repeats (IRs) region of 33,762 bp. The genome comprises of 135 genes, including 89 protein-coding genes, 38 tRNA genes, and 8 ribosomal RNA genes. The overall GC content was 36.96%. To confirm the phylogenetic relationship of P. polyphylla var. emeiensis, a maximum likelihood (ML) bootstraps analysis was performed based on 25 cp genomes. The sequences were aligned by MAFFT v7.307 (Katoh and Standley Citation2013; Jiang et al. Citation2020), and the phylogenetic tree was constructed by RAxML (Stamatakis Citation2014), with Dioscorea aspersa (NC 039807) and Dioscorea baya (NC 039850) as outgroups. Phylogenetic analysis showed that P. polyphylla var. emeiensis was closely related to P. fargesii and P. cronquistii (). The cp genome of P. polyphylla var. emeiensis will provide a useful resource for the conservation genetics of this species as well as for phylogenetic studies of Paris genus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank database at (https://www.ncbi.nlm.nih.gov) with the accession number is MT381945, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Additional information
Funding
References
- Duan BZ, Wang YP, Fang HL, Xiong C, Li XW, Wang P, Chen SL. 2018. Authenticity analyses of Rhizoma Paridis using barcoding coupled with high resolution melting (Bar-HRM) analysis to control its quality for medicinal plant product. Chin Med. 13:8
- Fu SZ, Li N, Liu Z, Gao WY, Man SL. 2012. Determination of seven kinds of steroidal saponins in plants of Paris L. from different habitats by HPLC. Chin Trad Herb Drugs. 12(43):2435–2437.
- Jiang Y, Wang J, Qian J, Xu L, Duan BZ. 2020. The complete chloroplast genome sequence of Oroxylum indicum (L.) Kurz (Bignoniaceae) and its phylogenetic analysis. Mitochondrial DNA B. 5(2):1429–1430.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu TX, Qian J, Du ZF, Wang J, Duan BZ. 2020. Complete genome and phylogenetic analysis of Agrimonia pilosa Ldb. Mitochondrial DNA B. 5(2):1435–1436.
- LIU Y, LUO D, YAO HUI, ZHANG X, YANG L, DUAN B. 2017. A new species of Paris (Melanthiaceae) from Yunnan, China. Phytotaxa. 326(4):297–300.
- Park JS, Choi YG, Yun N, Xi H, Min J, Kim YS, Oh S. 2019. The complete chloroplast genome sequence of Viburnum erosum (Adoxaceae). Mitochondrial DNA B. 4(2):3278–3279.
- Qian J, Du ZF, Jiang Y, Duan BZ. 2020. The complete chloroplast genome sequence of Althaea rosea (L.) Cavan. (Malvaceae) and its phylogenetic analysis. Mitochondrial DNA B. 5(2):1433–1434.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45 (W1):W6–W11.
- Yin HX, Zhang H, Xue D. 2007. Paris polyphylla var. emeiensis H. X. Yin, H. Zhang D. Xue, a new variety of Trilliaceae from Sichuan, China. Acta Phytotaxon Sin. 45(06):822–827.