Abstract
The Maccullochella peeli belongs to family Maccullochella, and is distributed in Australia and South America (mainly Argentina and Chile). In this paper, the complete mitochondrial genome of M. peeli was determined using next-generation sequencing. The whole mitogenome is a typical circular DNA molecule of 16,442 bp and contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a D-loop region, with the base composition of A 31.6%, G 14.3%, T 26.3%, and C 27.8%. Phylogenetic analysis showed that M. peeli was the nearest sister to Macquaria australasica. Our whole mitogenome presented here would be useful for further study of M. peeli.
The Maccullochella peeli belongs to the family Maccullochella and the species is distributed in Australia and South America (mainly Argentina and Chile) (Rowland Citation1983, Citation1989). Limited mitochondrial genome sequences have been reported from family Maccullochella. Here, we sequenced the complete mitochondrial genome of the adult fish, collected from Foshan in Guangdong, (latitude: 22°53′12.06′′N, longitude: 112°58′37.33′′E), Foshan Xinrong Fisheries Company, to provide reference information for further study on this species.
The typical specimen was deposited in the Jiangxi Agricultural University aquatic museum (M.peeli-001). Total genomic DNA was extracted from muscle tissue using the standard phenol-chloroform protocol (Barnett and Larson Citation2012). Then, the paired-end DNA library with an insert size of 400 bp was constructed and sequenced by Illumina X-ten with 150 bp in read length (Zhang et al. Citation2018). The de novo assembly of the mitochondrial genome was assembled by NOVOplasty (Dierckxsens et al. Citation2016).
The complete mitochondrial genome of M. peeli (GenBank Accession no. MN842722, 16,442 bp) contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a D-loop region. The whole base composition of the mitogenome is showed as follows: A 31.6%, G 14.3%, T 26.3%, and C 27.8%. The total length of the 13 protein-coding genes is 11,435 bp, all of which are encoded on the heavy strand, except for nad6, which was in the light strand.
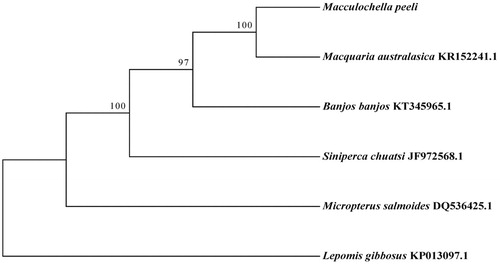
We performed a phylogenetic analysis of M. peeli species and Lepomis gibbosus outgroup species, Siniperca chuatsi, Micropterus salmoides, Banjos banjos, and Macquaria australasica based on 13 protein-coding genes sequences using maximum-likelihood method implemented in the RAxML (Silvestro and Michalak Citation2012). Macquaria australasicawas was the nearest sister to M. peeli (). The complete mitochondrial genome of M. peeli we determined would be useful in systematics and population genetics.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MN842722, reference number MN842722.
Additional information
Funding
References
- Barnett R, Larson G. 2012. A phenol-chloroform protocol for extracting DNA from ancient samples. Meth Mol Biol. 840:13–19.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Rowland SJ. 1983. Spawning of the Australian freshwater fish Murray cod, Maccullochella peeli (Mitchell), in earthen ponds. J Fish Biol. 23(5):525–534.
- Rowland SJ. 1989. Aspects of the history and fishery of the Murray cod, Maccullochella peeli (Mitchell) (Percichthyidae). Proc Linnean Soc New South Wales. 111(3):201–213.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Zhang Z, Zhang K, Chen S, Zhang Z, Zhang J, You X, Bian C, Xu J, Jia C, Qiang J, et al. 2018. Draft genome of the protandrous Chinese black porgy. Gigascience. 7(4):1–7.