Abstract
Chalcidoidea (Hymenoptera) are minute wasps which can attack immature and adult stages of virtually all insect orders. Here, we sequenced and annotated the mitochondrial genome (mitogenome) of Chalcidoidea sp. This mitogenome was 15,152 bp long and encoded 13 protein-coding genes (PCGs), 20 transfer RNA genes (tRNAs), and 2 ribosomal RNA unit genes (rRNAs). All 13 PCGs were initiated by the ATN (ATG, ATT, ATA, and ATC) codon. All PCGs terminate with the stop codons TAA or TAG except for nad4 which ended with the incomplete codon T−. Phylogenetic analysis showed that Chalcidoidea sp. got together with the species Encyrtus infelix and Eurytoma sp., and species in Chalcidoidea formed a sister group to other Cynipoidea and Proctotrupoidea species.
The superfamily Chalcidoidea is one of the most diverse groups of parasitic Hymenoptera. Most chalcid wasps are parasitoids that are successfully used as biological control agents of agricultural and ornamental pests, and they have tremendous importance in both natural and managed ecosystems (Gibson et al. Citation1999; Munro et al. Citation2011).
Specimens of Chalcidoidea sp. were collected from Baoji City, Shaanxi Province, China (33°51′N, 107°22′E, June 2019) and were stored in Entomological Museum of Anyang Institute of Technology (Accession number AIT-E-CHA06). After morphological identification, total genomic DNA was extracted from tissues using DNeasy DNA Extraction kit (Qiagen, Hilden, Germany). The mitogenome sequence of Chalcidoidea sp. was generated using Illumina HiSeq 2500 Sequencing System. In total, 6.9 G raw reads were obtained, quality-trimmed, and assembled using MITObim v 1.7 (Hahn et al. Citation2013). By comparison with the homologous sequences of other Chalcidoidea species from GenBank, the mitogenome of Chalcidoidea sp. was annotated using software GENEIOUS R8 (Biomatters Ltd., Auckland, New Zealand).
The nearly complete mitogenome of Chalcidoidea sp. is 15,152 bp (Genbank accession, MT419887). It contains 13 protein-coding genes (PCGs), 20 tRNA genes, 2 rRNA genes, and a partial non-coding AT-rich region. Mitogenome of Chalcidoidea sp. exhibit dramatic mitochondrial gene rearrangement which is common in Chalcidoidea species (Zhu et al. Citation2018; Tang et al. Citation2019; Xiong et al. Citation2019). The overall base composition of the mitogenome was estimated to be A 46.5%, T 37.3%, C 10.1%, and G 6.1%, with a high A + T content of 83.8%. All 13 PCGs of Chalcidoidea sp. have the conventional ATN start codons for invertebrate mitochondrial PCGs (six ATT, five ATG, and one each of ATA and ATC). Most of the PCGs terminate with the stop codon TAA or TAG, whereas nad4 end with the incomplete codon T. Two rRNA genes (rrnL and rrnS) located at trnL1/trnA and trnA/trnV, respectively. The lengths of rrnL and rrnS in Chalcidoidea sp. were 1301 and 776 bp, with the AT contents of 85.9 and 86.2%, respectively. Two tRNA genes (trnC and trnY) were not predicted in this mitogenome, and the other 20 tRNA genes vary from 64 bp (trnI and trnS1) to 74 bp (trnQ).
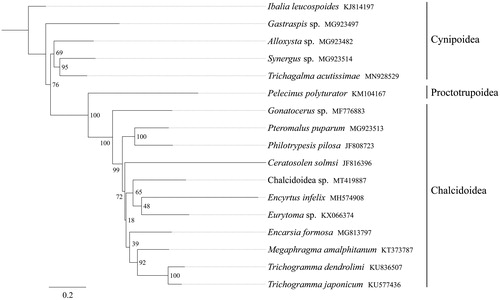
All 13 mitochondrial protein-coding genes sequences were extracted from the mitochondrial DNA sequences of 17 related taxa of Apocrita. A phylogenetic tree was constructed through raxmlGUI 1.5 (Silvestro and Michalak Citation2012). Results showed that the newly sequenced species Chalcidoidea sp. got together with the species Encyrtus infelix and Eurytoma sp., and all 11 Chalcidoidea species constituted one clade and formed a sister group to other Cynipoidea and Proctotrupoidea species. (). In conclusion, the mitogenome of Chalcidoidea sp. is sequenced in this study and can provide essential DNA molecular data for further phylogenetic and evolutionary analysis of Apocrita.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MT419887.
Additional information
Funding
References
- Gibson GAP, Heraty JM, Woolley JB. 1999. Phylogenetics and classification of Chalcidoidea and Mymarommatoidea — a review of current concepts (Hymenoptera, Apocrita). Zool Scripta. 28(1–2):87–124.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Munro JB, Heraty JM, Burks RA, Hawks DC, Mottern JL, Cruaud A, Rasplus J, Jansta P. 2011. A molecular phylogeny of the Chalcidoidea (Hymenoptera). PLoS One. 6(11):e27023.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers E. 12(4):335–337.
- Tang P, Zhu JC, Zheng BY, Wei SJ, Sharkey M, Chen XX, Vogler AP. 2019. Mitochondrial phylogenomics of the Hymenoptera. Mol Phylogenet Evol. 131:8–18.
- Xiong M, Zhou QS, Zhang YZ. 2019. The complete mitochondrial genome of Encyrtus infelix (Hymenoptera: Encyrtidae). Mitochondr DNA B. 4(1):114–115.
- Zhu JC, Tang P, Zheng BY, Wu Q, Wei SJ, Chen XX. 2018. The first two mitochondrial genomes of the family Aphelinidae with novel gene orders and phylogenetic implications. Int J Biol Macromol. 118(A):386–396.