Abstract
In this study, we sequenced and analyzed the complete mitochondrial genome of Drosophila busckii (Diptera: Drosophilidae). The mitogenome was 15,214 bp in length, containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes. The gene organization of D. busckii is identical to the ancestral gene arrangement found in most insects. All protein-coding genes started with ATN, except for cox2 and nad5, which used noncanonical codon TTG and GTG, respectively.
The fruit fly genus Drosophila (Diptera: Drosophilidae) is a diverse insect group with more than 1600 species worldwide (O’Grady and DeSalle Citation2018). Species of the genus Drosophila contain model organisms used in several biological research fields, and economically pests such as Drosophila suzukii (Kim et al. Citation2016). Nine subgenera were divided, including Dorsilopha, Drosophila, Sophophora, and so on. Drosophila busckii belongs to the subgenus Dorsilopha, in which only four species were recorded (O’Grady and DeSalle Citation2018). In this study, we report the mitochondrial genome of Drosophila busckii, representing the first mitogenome of the subgenus Dorsilopha.
Adults of D. busckii were collected in Nanjing (32.04° N and 118.89° E), Jiangsu province, China. The voucher specimen (DJNS) and its DNA were deposited in the Nanjing Agricultural University, Nanjing, China. Genomic DNA was extracted using Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China) following the manufacturer’s protocols. The DNA concentration was detected by Qubit 3.0 using Q33230 Qubit™ 1X dsDNA HS Assay Kit (ThermoFisher, Waltham, MA). The DNA library was sequenced on the Illumina Hi-Seq platform generating 150 bp paired-end reads. The mitogenome was assembled using NOVOPlasty v2.7.2 (Dierckxsens et al. Citation2017), and annotated via the software MitoZ (Meng et al. Citation2019).
The complete mitochondrial genomes of D. busckii was 15,214 bp in length (GenBank Accession No. MT429169). The mitogenome encodes the whole set of 37 genes (13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes) as observed in most insects. The gene organization of D. busckii is identical to that of the ancestral gene arrangement in most insects (Boore Citation1999).
The typical start codons ATN are assigned to 11 of all protein-coding genes PCGs. The genes cox1 and nad5 initiate with TTG and GTG, respectively. For 13 PCGs, four stop codons are found: T (cox2, nad1), TA (nad4, nad5), TAG (cytb) and TAA (other eight PCGs). The predicted secondary structures of all tRNA genes are typical cloverleaf except for trnS1 (AGN), lacking the dihydrouridine (DHU) stem. The nad4l–nad4 overlap was 1 bp in size, and not identical to that of atp8–atp6 (ATGATAA). The intergenic spacer between trnS2 (UCN) and nad1 was 18 bp in length.
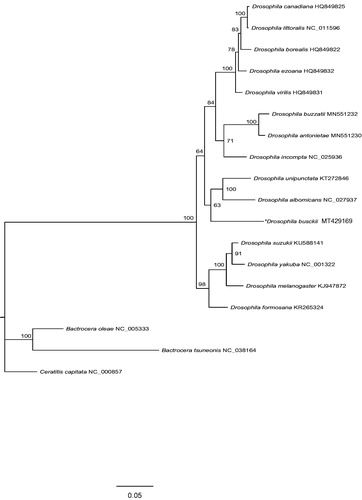
Each PCG was translated into an amino acid sequence, aligned with MAFFT v7.394 (Katoh and Standley Citation2013), and trimmed using trimAl v1.4.1 (Capella-Gutiérrez et al. Citation2009). The concatenated dataset of 13 PCGs were generated using FASconCAT-G v1.04 (Kück and Longo Citation2014). A maximum-likelihood tree () was inferred employing protein mixture model C60 with a bootstrap of 1000 replicates in the IQ-TREE (Nguyen et al. Citation2015). Three species of Tephritidae were chosen as outgroups. In the ML tree, the position of D. busckii, which clustered with Drosophila albomicans and Drosophila unipunctata, was in the clade of subgenus Drosophila, indicating the non-monophyletic subgenus Drosophila. The subgenus Sophophora represented by Drosophila melanogaster, Drosophila yakuba and Drosophila suzukii, was clustered with Drosophila formosana. The relationships between subgenus Dorsilopha and other fruit fly species groups need to be further investigated.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in figshare at http://10.6084/m9.figshare.12250331.
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim JS, Min MJ, Choi DS, Kim I. 2016. Complete mitochondrial genome of the spotted-wing drosophila, Drosophila suzukii (Diptera: Drosophilidae). Mitochondrial DNA Part B. 1(1):222–223.
- Kück P, Longo GC. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 11(1):81.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- O’Grady PM, DeSalle R. 2018. Phylogeny of the genus Drosophila. Genetics. 209(1):1–25.