Abstract
Picea is a phylogenetically complicate genus with great economic and ecological values. Here, we determined the whole complete chloroplast genome of Picea schrenkiana to provide genomic information for phylogenetic analysis of the genus. The plastome of P. schrenkiana is 124,060 bp in size and contains 114 genes, including 74 protein-coding genes, 36 tRNA genes, and four rRNA genes. The overall GC content is 38.7%. Unlike the typical plastome with a conserved quadripartite structure, loss of inverted repeat regions is found in the chloroplast genome. The phylogenetic tree shows that monophyly of P. schrenkiana is well supported.
Picea is a genus of about 35 species of coniferous evergreen trees widely distributed in the North Hemisphere, with great economic and ecological values. The monophyly of the genus has never been doubted, but infrageneric classification and interspecific relationships are quite controversial due to morphological convergence, reticulate evolution, and incomplete lineage sorting (Ran et al. Citation2006, Citation2015). Next-generation sequencing technology provides massive genome information for species discrimination and resolution to the complicated phylogenetic issues. Schrenk’s spruce (Picea schrenkiana), native to Tianshan mountains, is a major component of subalpine coniferous forest in central Asia. Its population size has sharply decreased for the global climate changes. In the present study, we characterized the complete chloroplast genome of P. schrenkiana and analyzed its phylogenetic position. Our results will supply effective data not only for plastome evolution, phylogenetic relationships of Picea species, but also for population genetics and conservation works in P. schrenkiana.
Genomic DNA was extracted from silica gel dried leaves of P. schrenkiana collected in Xinjiang, China (N43.89°, E88.14°). The voucher (2018LIU039) was deposited at the Evolutionary Botany Laboratory (EBL), Northwest University. Data treatments followed our previous study (Li et al. Citation2019), including data trimming, genome assembly, and annotation. The cp genome was annotated with Picea crassifolia (NC 032366) as a reference and has been deposited into GenBank with the accession number of MN871923.
We obtained a larger and more complete plastome sequence of P. schrenkiana than those previously reported (Sullivan et al. Citation2017). The plastome is 124,060 bp long and comprised of 74 protein-coding genes, 36 tRNA genes, and four rRNA genes. The overall GC content is 38.7%. Among the annotated genes, twelve contain a single intron and two (rps12 and ycf3) possess two introns. A typical cp genome of angiosperms has a conserved quadripartite structure with a large and small single copy region (LSC/SSC) separated by a pair of inverted repeat regions (IRs). By contrast, plastomes in Pinaceae species generally exhibit specific repeats (Wu et al. Citation2011). We examined the size variation of repeat sequences in eleven Picea species () and found that two copies of inverted repeats were generally located in ycf12 and its adjoined genes with the size range of 290–509 bp. Picea neoveitchii presented an additional pair of repeat sequences around the trnI-CAU and psbA gene. No IR-like sequence was detected in both P. schrenkiana and P. jezoensis. Consequently, Wu et al. (Citation2011) proposed that large repeat sequences other than highly reduced IRs played a critical role in evolution and recombination of gymnosperm plastomes (Sullivan et al. Citation2017).
Figure 1. The phylogenetic tree of eleven Picea species constructed by complete chloroplast genomes. The bootstrap values labeled beside the branches were based on 1000 replicates.
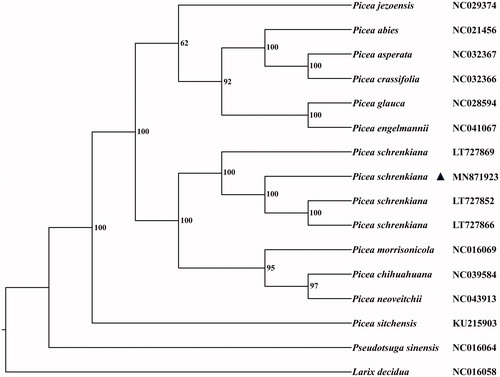
After alignment with MAFFT v7.310 (Katoh and Standley Citation2016), 14 plastomes of Picea were used to construct the Maximum-Likelihood (ML) tree with Pseudotsuga sinensis and Larix decidua as the outgroup. The procedure of tree construction was referred to Peng et al. (Citation2019). Our result shows that P. schrenkiana is clustered together with its congeners and interspecific relationships of Picea could be also clearly clarified in the ML tree ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are available on request from the corresponding author, or in NCBI at https://www.ncbi.nlm.nih.gov.
Additional information
Funding
References
- Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics. 32(13):1933–1942.
- Li JF, Yang Q, Liu ZL. 2019. The complete chloroplast genome sequence of Liparis japonica (Orchidaceae). Mitochondrial DNA B. 4(2):2405–2406.
- Peng FF, Chen L, Yang Q, Tian B, Liu ZL. 2019. The complete chloroplast genome of Dipelta yunnanensis (Caprifoliaceae), a vulnerable plant in China. Mitochondrial DNA B. 4(1):515–516.
- Ran JH, Shen TT, Liu WJ, Wang PP, Wang XQ. 2015. Mitochondrial introgression and complex biogeographic history of the genus Picea. Mol Phylogenet Evol. 93:63–76.
- Ran JH, Wei XX, Wang XQ. 2006. Molecular phylogeny and biogeography of Picea (Pinaceae): implications for phylogeographical studies using cytoplasmic haplotypes. Mol Phylogenet Evol. 41(2):405–419.
- Sullivan AR, Schiffthaler B, Thompson SL, Street NR, Wang XR. 2017. Interspecific plastome recombination reflects ancient reticulate evolution in Picea (Pinaceae). Mol Biol Evol. 34(7):1689–1701.
- Wu CS, Lin CP, Hsu CY, Wang RJ, Chaw SM. 2011. Comparative chloroplast genomes of Pinaceae: insights into the mechanism of diversified genomic organizations. Genome Biol Evol. 3:309–319.
