Abstract
We sequenced the mitochondrial genome of Daurian redstart Phoenicurus auroreus using the next-generation sequencing. The circular genome is 16,832 bp long, encoding 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a control region. The lengths of rrnL and rrnS were determined to be 1,597 and 984 bp, respectively. The phylogenetic analysis of 18 mitogenomes of Muscicapidae supports monophylies of all genera, including P. auroreus. The information would be useful for understanding the phylogeny and evolution of Muscicapidae.
The Daurian redstart (Phoenicurus auroreus) is a small passerine bird belongs to the family Muscicapidae, and is widely distributed in a large range of temperate Asia (BirdLife International Citation2020). The taxonomic status of P. auroreus, as well as some other lineages of the family, remain controversial (Tu et al. Citation2019; Chen et al. Citation2020). Here, we sequenced and assembled the complete mitochondrial genome of P. auroreus. The information would be helpful to infer the phylogenetic relationships of Muscicapidae.
The sample of P. auroreus, which was died of natural causes, were collected at Guyang, Baotou, Inner Mongolia, China (40°53′N, 110°13′E). The specimens were stored in the Museum of Baotou Teachers’ college (BA850127, Baotou Teachers’ college, China). Total genomic DNA was extracted using the DNeasy tissue kit (QIAGEN) following the manufacturer’s standard protocol. The library was constructed using an Illumina TruSeq Library Preparation kit and sequenced on an Illumina HiSeq 2500 platform. The raw data were trimmed using fastp (Chen et al. Citation2018) to remove adapters and low-quality reads. High-quality reads were assembled using MITObim v1.9.1 (Hahn et al. Citation2013), and annotated using MitoZ (Meng et al. Citation2019). The annotated mitogenome has been deposited in GenBank database under the accession number of MT366880.
The complete mitochondrial genome of P. auroreus is a typical circular DNA molecule with 16,832 bp in length, encoding 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a control region. the A–T content of P. auroreus mitogenome is 56.30%, which is the moderate of the known flycatcher mitogenomes. The major strand (J strand) carried most of the genes (12 PCGs and 14 tRNAs), while only nad6 had the remaining eight tRNAs which were on the minor strand (N strand). Most PCGs initiated with typical ATG start codons, except for cox1 initiating with GTG. The lengths of rrnL and rrnS were determined to be 1597 and 984 bp, respectively.
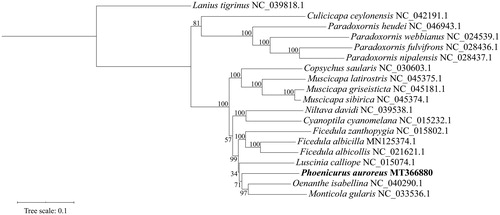
Eighteen complete mitogenomes of Muscicapidae, including P. auroreus, were used to infer the phylogenetic relationships among flycatchers based on the maximum likelihood (ML) analysis utilizing IQ-TREE 2 (Minh et al. Citation2019). The resulting tree supports monophylies of all genera, including P. auroreus in the present study (). The sequence data would be useful for elaboration of the phylogeny and evolution of Muscicapidae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the NCBI Genbank database at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT366880.
Additional information
Funding
References
- BirdLife International. 2020. Species factsheet: Phoenicurus auroreus; [accessed 2020 Apr 21]. http://www.birdlife.org.
- Chen P, Sun C, Liu B, Lu C, Chen Y. 2020. Complete mitochondrial genome of the reed parrotbill Paradoxornis heudei (Aves: Passeriformes: Muscicapidae). Mitochondrial DNA Part B. 5(2):1467–1468.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic acids Res. 41(13):e129–e129.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Minh BQ, Schmidt H, Chernomor O, Schrempf D, Woodhams M, Von Haeseler A, Lanfear R. 2019. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. BioRxiv. 849372.
- Tu H, Gao J, Jing J, Zhou C, Yue B, Zhang X. 2019. The complete mitochondrial genome of grey-headed canary-flycatcher (Culicicapa ceylonensis, Muscicapidae, Passeriformes). Mitochondrial DNA Part B. 4(1):1079–1080.