Abstract
Neope goschkevitschii and Lethe sicelis are endemic Satyrinae butterflies in mainland Japan, which belongs to the Palearctic realm. In this study, we determined the mitochondrial genomes of these two species. The total length of the mitochondrial genome was 15,286 bp and 15,196 bp for N. goschkevitschii and L. sicelis, respectively, and both mitochondrial genomes were extremely AT-rich. Phylogenetic analysis revealed each of these species was closely related to a member of the same genus, respectively.
Satyrinae is the largest subfamily of Nymphalidae, with 14 genera and 28 species distributed in Japan (Shirouzu Citation2006). There are six endemic species in Japan, four of which are found in the Ryukyu Archipelago, which are located in the Indomalayan realm. The other two species, Neope goschkevitschii and Lethe sicelis, are distributed in mainland Japan (Kyushu, Shikoku, Honshu, and Hokkaido), which are located in the Palearctic realm. Neope goschkevitschii is distributed from Hokkaido to Kyushu, and L. sicelis from Honshu to Kyushu and Shikoku. Both species inhabit forests in plains and low mountains, usually 2–3 broods per year; their larvae feed on members of Bambusoideae, such as Phyllostachys bambusoides and Sasa veitchii (Shirouzu Citation2006). However, the mitochondrial genomes of these two species have not yet been determined.
In this study, the complete mitochondrial genomes of N. goschkevitschii and L. sicelis were determined. Specimen of N. goschkevitschii was collected at N 35.93, E 139.93 on 14 August 2016, and that of L. sicelis at N 36.00, E 139.91 on 18 September 2017. The specimens and DNA of these specimens were stored in the National Museum of Nature and Science, Japan (NSMT-I-L37926 for N. goschkevitschii and NSMT-I-L38449 for L. sicelis). Total DNA was extracted from right middle leg using DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). A whole genome library was created using NEBNext Ultra II FS DNA Library Prep Kit for Illumina (NEB, Ipswich, MA) and then sequenced using Miseq V2 micro kit (Illumina, San Diego, CA). The total reads were assembled using NOVOplasty (Dierckxsens et al. Citation2017). The partial sequence of the control region and confirmation of the circularity were performed by PCR and Sanger sequencing. The mitochondrial genome was annotated using MITOS Webserver (Bernt et al. Citation2013).
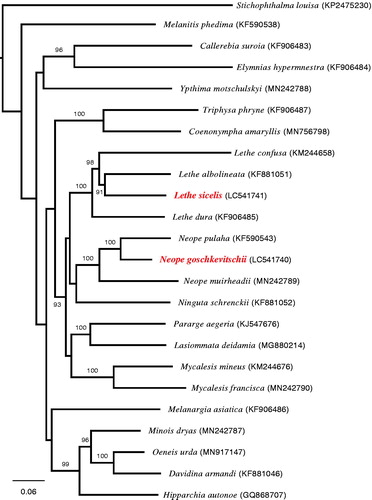
Total length of mitochondrial genomes of N. goschkevitschii (GenBank/DDBJ/EMBL Accession number LC541740) and L. sicelis (LC541741) were 15,286 bp and 15,239 bp, respectively. The mitochondrial genomes of both species were extremely AT-rich (the average A + T content was 79.8% for N. goschkevitschii, 79.5% for L. sicelis). Both of them were observed to possess 2 ribosomal RNA genes, 22 transfer RNA genes, 13 protein-coding genes (PCGs), and a control region (D-loop). Maximum likelihood analysis based on 13 PCGs using RAXML (Stamatakis Citation2014) with 24 Satyrinae species showed that three species in the Neope, including N. goschkevitschii, and four species in the Lethe, including L. sicelis, were monophyletic, respectively (). These mitogenomes are useful not only for phylogenetic relationships among species, but also for the estimation of biota formation in the Japanese archipelago.
Figure 1. Maximum likelihood phylogenetic tree of 24 Satyrinae butterflies based on 13 protein coding genes: Neope goschkevitschii (LC541740), Lethe sicelis (LC541741) (this study), Ninguta schrenckii (KF881052) (Fan et al. Citation2016), Stichophthalma louisa (KP247523) (Hou et al. unpublished), Melanargia asiatica (KF906486) (Huang et al. Citation2016), Hipparchia autonoe (GQ868707) (Kim et al. Citation2010), Lasiommata deidamia (MG880214) (Li et al. unpublushed), Lethe confusa (KM244658), Mycalesis mineus (KM244676) (Tang et al. Citation2014), Pararge aegeria (KJ547676) (Teixeira Citation2016), Melanitis phedima (KF590538) (Wu et al. Citation2014), Neope pulaha (KF590543) (Wu et al. Citation2014), Lethe albolineata (KF881051), Davidina armandi (KF881046) (Xu et al. unpublished), Ypthima motschulskyi (MN242788), Neope muirheadii (MN242789), Mycalesis francisca (MN242790), Minois dryas (MN242787) (Yang et al. Citation2020), Triphysa phryne (KF906487) (Zhang et al. Citation2016), Callerebia suroia (KF906483), Elymnias hypermnestra (KF906484), Lethe dura (KF906485) (Zhang et al. unpublished), Oeneis urda (MN917147) (Zhou et al. Citation2020), Coenonympha amaryllis (MN756798) (Zhou unpublished). The number beside each node indicate bootstrap values in percentage based on 1000 replications.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the GenBank (https://www.ncbi.nlm.nih.gov/nucleotide/) or DDBJ (http://getentry.ddbj.nig.ac.jp/top-e.html), accession numbers of all data are listed in this article.
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fan C, Xu C, Li J, Lei Y, Gao Y, Xu C, Wang R. 2016. Complete mitochondrial genome of a satyrid butterfly, Ninguta schrenkii (Lepidoptera: Nymphalidae). Mitochondrial DNA A. 27(1):80–81.
- Huang D, Hao J, Zhang W, Su T, Wang Y, Xu X. 2016. The complete mitochondrial genome of Melanargia asiatica (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA A. 27(2):806–808.
- Kim MJ, Wan X, Kim K, Hwang JS, Kim I. 2010. Complete nucleotide sequence and organization of the mitogenome of endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Afr J Biotechnol. 9:735–754.
- Shirouzu T. 2006. The butterflies of Japan in color. Tokyo, Japan: Gakken Holdings.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes – a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42(22):e166.
- Teixeira C. 2016. The complete mitochondrial genome of Parage aegeria (Insecta: Lepidoptera: Papilionidae). Mitochondrial DNA A. 27:551–552.
- Wu LW, Lin LH, Lees DC, Hsu YF. 2014. Mitogenomic sequences effectively recover relationships within brush-footed butterflies (Lepidoptera: Nymphalidae). BMC Genomics. 15:468.
- Yang M, Song L, Zhou L, Shi Y, Song N, Zhang Y. 2020. Mitochondrial genomes of four satyrine butterflies and phylogenetic relationships of the family Nymphalidae (Lepidoptera: Papilionoidea). Int J Biol Macromol. 145:272–281.
- Zhang W, Gan S, Zuo N, Chen C, Wang Y, Hao J. 2016. The complete mitochondrial genome of Triphysa phryne (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA A. 27(1):474–475.
- Zhou Y, Liang Z, Wang S, Zhong H, Wang N, Liang B. 2020. A mitogenomic phylogeny of satyrid butterflies and complete mitochondrial genome of Oeneis urda (Lepidoptera: Nymphalidae: Satyrinae). Mitochondrial DNA B. 5(2):1344–1345.
