Abstract
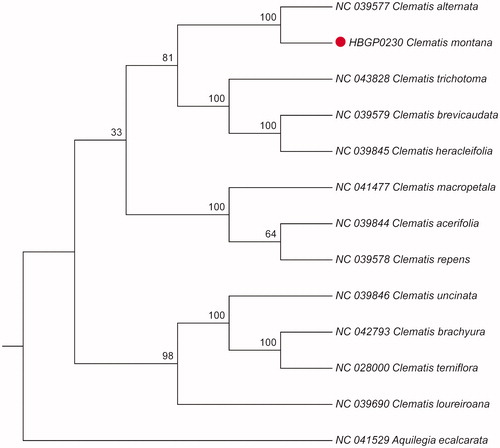
Clematis montana is a medicinal plant commonly used in southwest of China. The complete chloroplast (cp) genome sequence of C. montana was sequenced using the Illumina Hiseq 4000 platform. The cp genome of C. montana was 159,523 bp in length with 37.98% overall GC content. This circular molecule had a typical quadripartite structure containing a large single-copy (LSC) region of 79,385 bp, a small single-copy (SSC) region of 18,092 bp, and two inverted repeat (IR) regions of 31,023 bp. The cp genome contained 135 genes, including 91 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. Phylogenetic analysis based on whole cp genome sequences showed that C. montana was closest to C. alternata.
Clematis montana Buch.-Ham. belongs to the family of Ranunculaceae, which woody climber distributed in temperate Himalaya up to an altitude of 4000 m (Singhal et al. Citation2010). The dry stem of C. montana was used as traditional Chinese Medicine for the treatment of inflammatory conditions, such as rheumatism, urinary tract infection, and so on (Chawla et al. Citation2012; Bhatt et al. Citation2016). Previous studies mainly focused on describing its chemical compositions and their pharmacological activities. In this study, we reported the complete cp genome and phylogenetic analysis of C. montana for the first time, which will contribute to further studies on its genetic research and resource utilization.
Fresh leaves of C. montana were sampled from the Nanjing Botanical Garden Men. Sun Yat-Sen, Institute of Botany Jiangsu Province and Chinese Academy of Sciences (Nanjing, China, N32°03′6.64″, E118°49′39.54″). The voucher specimen was deposited in the herbarium of Institute of Chinese Meteria Medica, China Academy of Chinese Medical Sciences (accession number: HBGP0230_NJ). Genomic DNA was extracted using the DNeasy plant mini kit (Qiagen). The whole-genome sequencing was conducted with 350 bp pair-end reads on the Illumina NovaSeq system (Illumina, San Diego, CA). In total, 4.02 Gb of raw data (26,813,858 reads) were obtained. De novo genome assembly and annotation were conducted by NOVOPlasty (Dierckxsens et al. Citation2017) and GeSeq (Tillich et al. Citation2017), respectively. The annotated cp genome was deposited in the GenBank (accession number: MT292622).
The result showed that the cp genome of C. montana was 159,523 bp in length, with a large single-copy region (LSC) of 79,385 bp, a small single-copy region (SSC) of 18,092 bp, and a pair of inverted repeat (IR) regions of 31,023 bp. A total of 135 genes were annotated, including 91 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The GC content of the cp genome is 37.98%. To identify the phylogenetic position of C. montana, alignment was performed on the 13 cp genome sequences using MAFFT v7.309 (Katoh and Standley Citation2013). The maximum likelihood (ML) bootstrap analysis with 1000 replicates was performed using RaxML v8.2.12 (Stamatakis Citation2014). The phylogenetic tree showed that C. montana was closely related to C. alternata (). The cp genome sequence of C. montana in this study might provide important information on phylogenetic and evolutionary studies in Ranunculaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The annotated cp genome has been deposited in GenBank under accession number [MT292622].
The data that support the findings of this study are openly available in Genbank at [https://www.ncbi.nlm.nih.gov/nuccore/], reference number [MT292622].
Additional information
Funding
References
- Bhatt H, Saklani S, Upadhayay K. 2016. Prelimanary phytochemical, phytochemical, antioxidant and antimicrobial studies of Clematis montana leaves. Int J Curr Pharm Sci. 8(4):33.
- Chawla R, Kumar S, Sharma A. 2012. The genus Clematis (Ranunculaceae): chemical and pharmacological perspectives. J Ethnopharmacol. 143(1):116–150.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Singhal VK, Kaur S, Kumar P. 2010. Aberrant male meiosis, pollen sterility and variable sized pollen grains in Clematis montana Buch.-Ham. ex DC. from Dalhousie hills, Himachal Pradesh. Cytologia. 75(1):31–36.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–w11.