Abstract
The complete mitochondrial genome was sequenced from the marine water flea Diaphanosoma celebensis. The sequenced mitochondrial genome size was 17,060 bp, possessing identical gene order of 13 protein-coding genes (PGCs) to those of the congeneric freshwater species Diaphanosoma dubium in the genus Diaphanosoma. The mitochondrial genome of D. celebensis had 13 PGCs, two rRNAs, and 22 tRNAs. Of 13 PGCs, three genes (CO3, ND3, and ND4) had incomplete stop codons. Furthermore, the stop codons of the remaining ten PGCs were TAA (for CO1, ATP8, ATP6, ND5, ND6, and ND1) and TAG (for NL4L, Cytb, and ND2). The second and third base composition of codon on 9 PCGs on the L strand in D. celebensis mitogenome showed an anti-G bias (11.0% and 15.0%), respectively.
To date, more than 20 species are retrieved in the genus Diaphanosoma (http://v3.boldsystems.org/index.php/TaxBrowser_Taxonpage?taxid=5262), whereas Korovchinsky (Citation1989) and Korovchinsky (Citation1993) were firstly to redescribe Diaphanosoma celebensis in tropical Asia. Also, D. celebensis has been used in aquaculture (Segawa and Yang Citation1987), developmental biology (In et al. Citation2019), environmental toxicology (Kim et al. Citation2018), and other aspects, as one of the non-model organisms in the marine and brackishwater environment. In genus Diaphanosoma, the complete mitochondrial genome of Diaphanosoma dubium has been firstly reported (Liu et al. Citation2017), but no further studies on the complete mitochondrial genome have been reported from other Diaphanosoma spp. In this paper, we report the complete mitochondrial genome of the marine water flea D. celebensis as a life barcode that could be applicable as a potential non-model organism for multipurpose studies in the laboratory condition studies.
The adult D. celebensis were sampled in the estuarine area in Malaysia and maintained at the Laboratory of Professor Atsushi Hagiwara, Nagasaki University in Japan since August 1988 (kindly provided by Dr. Won Tack Yang, The Marine Biomedical Institute, The University of Texas Medical Branch). The type was deposited in the ichthyological collection of the Faculty of Fisheries, Nagasaki University (FFNU) under the accession no. FFNU-Cr-00394. We sequenced the whole genome of the marine water flea D. celebensis from whole body genomic DNA with Oxford Nanopore Technologies (Oxford UK). De novo assembly was performed with Nanopore sequences using flye V2.7 (https://github.com/fenderglass/Flye) and Medaka (https://github.com/nanoporetech/medaka) to correct errors in nanopore sequences and create a consensus nanopore sequences. Illumina reads including 500 bp paired-end sequencing were mapped to nanopore assembly to correct and polish the consensus sequences using Pilon (Walker et al. Citation2014) (https://github.com/broadinstitute/ pilon/wiki). Of the assembled 192 D. celebensis scaffolds (total 100,388,500 bp; N50 = 2.56 Mb; BUSCO value for eukaryota 96.7%), a single scaffold was mapped to the mitochondrial DNA of Diaphanosoma dubium (GenBank accession no. NC_037488). To obtain the complete mitochondrial genome of D. celebensis, we employed minimpa2 V2.17 (https://github.com/lh3/minimap2) and re-assembled it with canu V1.7 (https://github.com/marbl/canu).
The total length of the complete mitochondrial genome of D. celebensis is 17,060 bp (GenBank accession no. MT356995). The mitochondrial genome of D. celebensis contained 13 protein-coding genes (PGCs), two rRNAs, and 22 tRNAs. The direction of 13 PGCs and two rRNA genes of D. celebensis was identical to those of a congeneric freshwater water flea species D. dubium (Liu et al. Citation2017). Of 13 PCGs in D. celebensis mitochondrial genome, three genes (CO3, ND3, and ND4) had incomplete stop codons. The mitochondrial genome base composition of 13 PCGs was 35.6% for A, 32.4% for T, 13.6% for G and 18.4% for C. The A + T base composition (68.0%) was higher than G + C base composition (32.0%). The second and third base composition of codon on 9 PCGs on the L strand in D. celebensis mitogenome shows an anti-G bias (11.0% and 15.0%), respectively. The most commonly found start codon in D. celebensis was ATG but some genes used ATT (CO1, ND3, and ND6 genes) and ATA (ND4 gene), respectively.
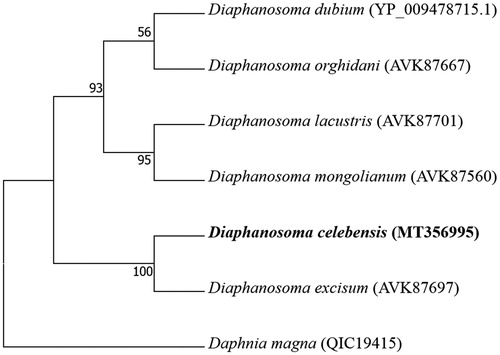
The placement of D. celebensis among six Diaphanosoma species with mitochondrial cytochrome oxidase 1 gene with outgroup (Daphnia magna) is shown in . Among six Diaphanosoma species, D. celebensis was the closest species to D. excisum, validated by high clusterization, compared to other four Diaphanosoma species.
Figure 1. Phylogenetic analysis. We conducted a comparison of the mitochondrial cytochrome oxidase 1 (CO1) gene of six species in the genus Diaphanosoma. The mitochondrial CO1 gene was aligned by ClustalW. Maximum-likelihood analysis was performed by Mega software (ver. 10.0.1) with LG + G + I model. The rapid bootstrap analysis was conducted with 1000 replications with 48 threads running in parallel. The cladoceran Daphnia magna served as an outgroup. Ln = −2021.49.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are available from the corresponding author, J.-S.L, upon reasonable request.
Additional information
Funding
References
- In S, Yoon HW, Yoo JW, Cho H, Kim R-O, Lee Y-M. 2019. Acute toxicity of bisphenol A and its structural analogues and transcriptional modulation of the ecdysone-mediated pathway in the brackish water flea Diaphanosoma celebensis. Ecotoxicol Environ Saf. 179:310–317.
- Kim B-M, Kang S, Kim R-O, Jung J-H, Lee K-W, Rhee J-S, Lee Y-M. 2018. De novo transcriptome assembly of brackish water flea Diaphanosoma celebensis based on short-term cadmium and benzo[a]pyrene exposure experiments. Hereditas. 155:36.
- Korovchinsky NM. 1989. Redescription of Diaphanosoma celebensis Stingelin, 1900 (Crustacea, Cladocera). Hydrobiologia. 184(1–2):7–22.
- Korovchinsky NM. 1993. New records of Diaphanosoma celebensis Stingelin 1900 (Crustacea: Daphniiformes: Sididae) in tropical Asia with remarks on the morphological variability and biology of the species. Rev Hydrobiol Trop. 26:119–125.
- Liu P, Xu S, Huang Q, Dumont HJ, Lin Q, Han B-P. 2017. The mitochondrial genome of Diaphanosoma dubium with comparison with Daphnia magna. Mitochondrial DNA B. 2(2):926–927.
- Segawa S, Yang W. 1987. Reproduction of an estuarine Diaphanosoma aspinosum (Branchiopoda: Cladocera) under different salinities. Bull Plankton Soc Jpn. 34:43–51.
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLOS One. 9(11):e112963.
