Abstract
Tree fern Alsophila spinulosa is an endangered relic plant in the world. It is currently on the International Union for Conservation of Nature (IUCN) red list of threatened species. In this study, we first assembled the complete chloroplast (cp) genome of A. spinulosa by Illumina paired-end reads data. The whole genome was 156,661 bp, consisting of a pair of inverted repeats of 24,364 bp, large single copy region and a small single copy region (70,352 and 21,624 bp in length, respectively). The cp genome contained 133 genes, including 92 protein-coding genes, 33 trRNA genes, and eight rRNA genes. The overall GC content of the whole genome was 40.4%. A neighbour-joining phylogenetic analysis demonstrated a close relationship between A. spinulosa and Cystoathyrium chinense Ching.
Alsophila spinulosa is the only relict tree fems since the Jurassic Period, which has been endangered by habitat fragmentation (Tryon and Tryon Citation1982). The community of A. spinulosa has been restored with the increase of conservation intensity, but recent studies have found that most Cyatheaceae communities are experiencing decline and seedling reduction, some scholars also believe that the appearance of this phenomenon may be related to human activities (Kiran et al. Citation2012).
In this study, A. spinulosa was sampled from the Endangered Species Reserve of Jilin Agricultural University Changchun County, China (125°19′E, 43°43′N). A voucher specimen (JN20200501) was deposited in the Herbarium of the Plant Biotechnology Center of Jilin Agricultural University, Changchun, China.
The present study is the first time to assemble and characterize the complete chloroplast genome for A. spinulosa (GenBank: NC_012818.1) from hight-hrough-put sequencing data. The existing chloroplast Genome sequence of ginkgo biloba was downloaded from the National Center for Biotechnology Information’s Organelle Genome Resources database (nc_016986.1) as the reference sequence, and the chloroplast Genome of A. spinulosa was assembled using SPAdes v3.6.0 software (Bankevich et al. Citation2012). The default setting of parameters was adopted. Sequence annotation first confirmed the availability and boundary of genes by blastn comparison directly through the protein-coding sequence of the proximal species. Then, the genes in the chloroplast genome were annotated by online tool DOGMA (http://dogma.ccbb.utexas.edu/) with default parameters, and the genes were functionally annotated by combining with NR (http://www.ncbi.nlm.nih.gov/) database (Lohse et al. Citation2013). TRNA was annotated using the trnascan-se online site. Using RNAmmer 1.2 Server (http://www.cbs.dtu.dk/services/RNAmmer/) rRNA for comments. The chloroplast genome of ginkgo biloba was mapped using OGDRAW (http://OGDRAW.Mpimp-golm.mpg.DE/cgi-bin/OGDRAW.Pl) software (Asaf et al. Citation2017).
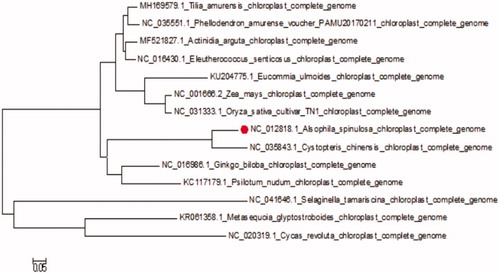
The complete cp-DNA of A. spinulosa was a circular molecule 156,661 bp in length, comprising a large single copy (LSC) region of 70,352 bp and a small single copy (SSC) region of 21,624 bp, separated by two inverted repeat regions (IRs) of 24,364 bp. It contained 133genes, including 92 protein-coding genes, 8 ribosomal RNA genes, and 33 tRNA genes. The phylogenetic tree reveals all the species of Pteridophyta formed a monophyletic clade with high-resolution value and Cystoathyrium chinense Ching is most related to A. spinulosa ().
Acknowledgements
Thanks to Gao, L., Yi, X., Yang, Y.X., Su, Y.J. and Wang, T. for the research provided a theoretical basis for my previous data. (Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes.)
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statements
The data that support the findings of this study are openly available in figshare at 10.6084/m9.figshare.12250268.
Additional information
Funding
References
- Asaf S, Waqas M, Khan AL, Khan MA, Kang SM, Imran QM, Shahzad R, Bilal S, Yun BW, Lee IJ. 2017. The complete chloroplast genome of wild rice (Oryza minuta) and its comparison to related species. Front Plant Sci. 8:304.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19 (5):455–477.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Kiran PM, Raju AV, Rao BG. 2012. Investigation of hepatoprotective activity of Cyathea gigantea (Wall. ex. Hook.) leaves against paracetamol-induced hepatotoxicity in rats. Asian Pac J Trop Biomed. 2(5):352–356.
- Tryon RM, Tryon AF. 1982. Ferns and allied plants: with special reference to tropical America. New York: Springer-Verlag; p. 138–212.