Abstract
Syringa oblata Lindl. is a popular ornamental shrub with aroma compounds. Here, we sequenced and assembled the complete chloroplast genome of S. oblata. The complete chloroplast genome of S. oblata is 155,648 bp in length, containing a pair of inverted repeated (IRa and IRb) region of 25,732 bp that are separated by a large single copy (LSC) region of 86,247 bp, and a small single copy (SSC) region of 17,937 bp. A total of 132 functional genes were annotated, including 88 protein-coding genes, 36 tRNA genes, and eight rRNA genes. The Neighbour-joining phylogenetic tree based on complete chloroplast genomes suggested that S. oblata is most closely related to S. vulgaris.
Syringa (Lilac) is among the popular ornamental bushes and is widely cultivated in the northern hemisphere. Flowers of Syringa oblata Lindl. bloom from April to May in China. This flower releases the aroma compounds, which are very pleasant to the human sensory system (Li et al. Citation2006). Chloroplast genome sequences are the most important source of genetic markers to study the distribution of paternal genes and paternally based molecular phylogenetic relationships (Shaw et al. Citation2007). In this study, the complete chloroplast genome (accession number: MT025818) of S. oblata was de novo sequenced based on new generation sequencing (NGS) technology.
The samples of S. oblata were collected from Xining Botanical Garden, Xining, China (36.60°N, 101.76°E). The experiment and analysis scheme refers to Wang et al. (Citation2019). The experiment and analysis scheme also refer to Wang et al. (Citation2019). Total DNA of S. oblata was extracted from the dried, young leaves (about 0.3 g) with a modified CTAB method (Doyle and Doyle Citation1987). The voucher specimen (Specimen Accession number: WJL2019035) was kept in Herbarium of the Northwest Institute of Plateau Biology, Chinese Academy of Sciences (HNWP). Genome sequencing was performed using the Illumina HiSeq Platform (Illumina, San Diego, CA) at Genepioneer Biotechnologies Inc., Nanjing, China. Approximately 6.13 GB of clean data was yielded. The trimmed reads were mainly assembled by SPAdes (Bankevich et al. Citation2012). The assembled genome was annotated using CpGAVAS (Liu et al. Citation2012).
The complete chloroplast genome of S. oblata is 155,648 bp in length, containing a pair of inverted repeated (IRa and IRb) region of 25,732 bp that are separated by a large single copy (LSC) region of 86,247 bp, and a small single copy (SSC) region of 17,937 bp. A total of 132 functional genes were annotated, including 88 protein-coding genes, 36 tRNA genes, and eight rRNA genes. The protein-coding genes, tRNA genes, and rRNA genes account for 66.67%, 27.27%, and 6.06% of all annotated functional genes, respectively.
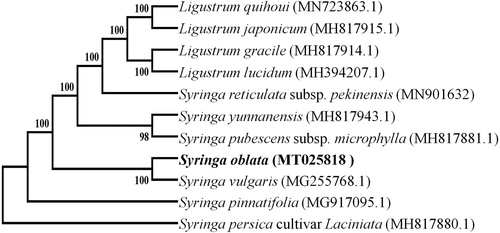
Phylogenetic relationships of S. oblata and its closely related species were resolved by means of Neighbour-joining (NJ). The alignment was conducted using MAFFT (Katoh and Standley Citation2013; online version: https://mafft.cbrc.jp/alignment/server/). The Neighbour-Joining (NJ) tree was constructed using the MEGA7 package (Kumar et al. Citation2016) with 1000 bootstrap repetitions. The NJ phylogenetic tree suggested that S. oblata is most closely related to S. vulgaris ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in Genbank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT025818.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li ZG, Lee MR, Shen DL. 2006. Analysis of volatile compounds emitted from fresh Syringa oblata flowers in different florescence by headspace solid-phase microextraction-gas chromatography-mass spectrometry. Anal Chim Acta. 576(1):43–49.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Shaw J, Lickey EB, Schilling EE, Small RL. 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot. 94(3):275–288.
- Wang J, Cao Q, He C, Ma Y, Li Y, Liu J, Zhang F. 2019. Complete chloroplast genome of Exacum affine (Gentianaceae): the first plastome of the tribe Exaceae in the family Gentianaceae. Mitochondrial DNA Part B. 4(2):3529–3530.