Abstract
The complete mitochondrial genome of the Omphalius rusticus has been determined. The complete genome is 18,067 bp and contained 13 protein-coding genes, two rRNA genes and 22 tRNA genes. The overall base composition is 33.48% (A), 33.52% (T), 15.58% (G) and 17.42% (C). The all start codon for 13 protein-coding genes is ATG and the most common termination codon is TAA. The phylogenetic tree showed that O. rusticus is most closely related to the Tectus pyramis. We suggest that this result will further supplement the genome information in mitochondria of the family Tegulidae and facilitate the study on population genetics.
Omphalius rusticus belongs to the family Tegulidae. It is widely distributed along the coast of China, South Korea (Sato-Okoshi et al. Citation2012), Japan (Fuji and Nomura Citation1990), and other western Pacific regions. The length of adult shells is between 10 and 20 mm. It is an edible perennial gastropod that can live for 5 years or more, and it is herbivorous. O. rusticus is dioecious and oviparous, and the spawning stage took place from July to September (Lee Citation2001). O. rusticus has been developed as a kind of sea food because of its high protein and low fat. To date, however, less research is relative to the sequence of the mitochondrial DNA genes of O. rusticus.
Herein, it is the first time to determine the complete mitochondrial genome of O. rusticus. This study will help to understand the phylogenetic status of genus Omphalius among the Class Gastropoda and Phylum Mollusca. The specimen of O. rusticus was collected in Penglai, Shandong Province, China (120.7°E, 37.8°N) and identified by morphology and deposited in Zhejiang Ocean University (accession number: OR20181021). The total DNA extraction utilized the salting-out method (Aljanabi and Martinez Citation1997) with the muscle. Then, total genomic DNA was diluted to a final concentration of 60–80 ng/µl in 1× TE buffer and stored at 4 °C. The genomic DNA was prepared in 400 bp paired-end libraries. The Illumina HiSeq X Ten platform was used to perform the high-throughput sequence. All the data were available and enumerated to the Microsoft oneDrive database (https://1drv.ms/w/s!ArF1Al5lLW_Vctd6wLfMq_cTw-E?e=5i4rMT).
The length of the complete mitochondrial genome of O. rusticus is 18,067 bp with the GenBank accession no. MT366560. The complete mitochondrial genome has 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA (tRNA) genes. The mitochondrial base composition is A 33.48%, T 33.52%, G 15.58%, and C 17.42%, with an obvious (A + T) % > (G + C) %. In 13 protein-coding genes, all of them start with ATG. For the stop codon, atp8 ends with TAG and nad3 ends with TGA, other 11 genes end with TAA. The 16S rRNA is 1560 bp between the tRNALeu1 and tRNAVal, and the 12S rRNA is 1005 bp between the tRNAVal and tRNAMet.
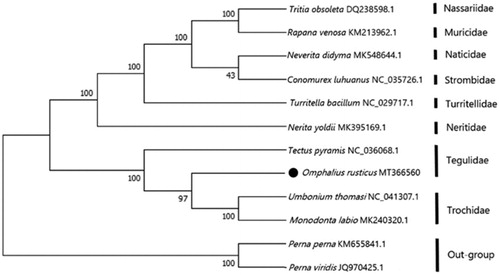
The phylogenetic relationship is estimated using the Neighbour-joining method (Schull Citation1979) in the program Phylip (Felsenstein Citation1989) based on 13 protein-coding genes of O. rusticus and other 11 species (). It is showed that the phylogenetic relationship of O. rusticus is very close to the Tectus pyramis. Meanwhile, the phylogenetic relationship of O. rusticus is far away from Perna perna and Perna viridis. We expect the present results will further supplement the genome information in mitochondria of the family Tegulidae and facilitate the study on the taxonomy, population genetic structure, and phylogenetic relationships.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in Microsoft OneDrive at https://1drv.ms/w/s!ArF1Al5lLW_Vctd6wLfMq_cTw-E?e=5i4rMT; and in GenBank, reference number: MT366560.
Additional information
Funding
References
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25(22):4692–4693.
- Felsenstein J. 1989. PHYLIP-Phylogeny inference package (Version 3.2). Cladistics. 5:164–166.
- Fuji A, Nomura H. 1990. Community structure of the rocky shore macrobenthos in southern Hokkaido. Mar Biol. 107(3):471–477.
- Lee JH. 2001. Gonadal development and reproductive cycle of the top shell, Omphalius rusticus (Gastropoda: Trochidae). J Korean J Biol Sci. 5(1):37–44.
- Sato-Okoshi W, Okoshi K, Koh B-S, Kim Y-H, Hong J-S. 2012. Polydorid species (Polychaeta: Spionidae) associated with commercially important mollusk shells in Korean waters. J Aquacult. 350–353:82–90.
- Schull WJ. 1979. Genetic structure of human populations. J Toxicol Environ Health. 5(1):17–25.