Abstract
Osedaxrubiplumus(Annelida, Siboglinidae)uses heterotrophic bacteria to feed onvertebrate carcasses and is currently found in the Pacific, Antarctic and Indian Ocean.Here, we report its nearly complete mitochondrial genomes assembled for 2 individuals, one from the East Pacific and the other from the Southwest Indian Ocean. Recoveredmitogenomes were 15591 and 15972 bp in length, with both consisting of 37 typical metazoan mitochondrial genes. All genes were transcribed from the same strand, and arranged in the same order as the other siboglinids, revealing conserved gene arrangement withinSiboglinidae. Phylogeneticanalysis of 13 protein coding genes confirms the placement of Osedaxsister to the Vestimentifera+Sclerolinum clade.
The bone-eating worms, Osedax spp., are specialists thriving in chemosynthetic ecosystems formed by whale falls and other vertebrate carcassesthrough out the world’soceans (Amon et al. Citation2014; Rouse et al. Citation2018; Zhou et al. Citation2020). They aresiboglinidannelidsand recent analyses (Li et al. Citation2015, Citation2017) placed them as sister to the Vestimentiferan + Sclerolinum clade.Although the phylogenetic position is well understood, the mitogenome has not been well explored in Osedax(Li et al. Citation2015) and its gene arrangement is still unknown. Interestingly, all sequenced siboglinidmtDNA shared the same gene arrangement (Li et al. Citation2015). Thus,a wider taxon sampling is needed to better explore the mitogenomein Osedax.
Specimens of O. rubiplumuswere collected fromEast Pacific margin (43°54.52′N; 125°10.29′W, 1560 m) and Southwest Indian Ridge (49°38.685′E, 37°47.013′S, 2908 m), which have been deposited in Auburn University (vouch number: AUMNH 46876) and the Repository of the Second Institute of Oceanography, MNR (vouch number: RSIO49bone_ind) respectively. One individual from each sampling site was used forgenomic DNA extraction, sequencing, assembling and gene annotationfollowing the methods described in Li et al. (Citation2015) and Zhou et al. (Citation2019) respectively. A maximum likelihood (ML) analysis based on concatenated alignments of the amino acid sequences of the 13 PCGs was conductedin IQtree 1.6.10 (Trifinopoulos et al. Citation2016) with substitution model for each individual gene or partition determined by the program automatically.
Twomitogenomes of Osedaxrubiplumus (GenBank accession numbers: MT108936 and MT108937, 15591 and 15972 bp in length respectively) contains 13 PCGs, 2rRNA genes and 22 tRNA genes. Consistent with other siboglinidmitogenomes, the 13 PCGs use ATG as the start codon, and a combination of TAA, TAG and T as stop codon. All genes are transcribed from the same strand, and arranged in the sameorder asother siboglinids, suggesting conserved gene arrangement in the family (Li et al. Citation2015).
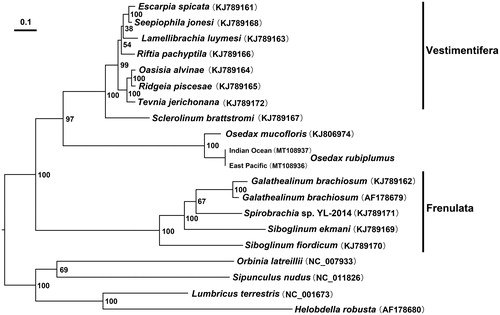
Previous study revealed four major lineages in Siboglinidae: Vestimentifera, Frenulata, Sclerolinum, and Osedax (Rouse et al., Citation2004; Halanych Citation2005). Using the 13 PCGs of Osedaxmucofloris mitochondrion, Li et al. (Citation2015) Li et al. (Citation2017) suggested that Osedax is genetically closer to the Vestimentifera + Sclerolinum claderather than to Frenulata. The maximum likelihoodanalysis in the present study robustly supports the sister relationship between the Osedax and Vestimentifera + Sclerolinum clade ().
Disclosure statement
The authors declare no conflict of interest.The authors alone are responsible for the content and writing of the paper.
Data availability statement
All sequencesgenerated or used in the present study are deposited in NCBI GenBank (https://www.ncbi.nlm.nih.gov) and the accession numbers are detailed in .
Additional information
Funding
References
- Amon DJ, Wiklund H, Dahlgren TG, Copley JT, Smith CR, Jamieson AJ, Glover AG. 2014. Molecular taxonomy of Osedax (Annelida: Siboglinidae) in the Southern Ocean. Zool Scr. 43(4):405–413.
- Halanych KM. 2005. Molecular phylogeny of siboglinid annelids (a.k.a. pogonophorans): a review. Hydrobiologia. 535–536(1):297–307.
- Li YN, Kocot KM, Schander C, Santos SR, Thornhill DJ, Halanych KM. 2015. Mitogenomics reveals phylogeny and repeated motifs in control regions of the deep-sea family Siboglinidae (Annelida). Mol Phylogenet Evol. 85(2015):221–229.
- Li YN, Kocot KM, Whelan NV, Santos SR, Waits DS, Thornhill DJ, Halanych KM. 2017. Phylogenomics of tubeworms (Siboglinidae, Annelida) and comparative performance of reconstruction methods. Zool Scr. 46(2):200–213.
- Rouse GW, Goffredi SK, Johnson SB, Vrijenhoek RC. 2018. An inordinate fondness for Osedax (Siboglinidae: Annelida): fourteen new species of bone worms from California. Zootaxa. 4377(4):451–489.
- Rouse GW, Goffredi SK, Vrijenhoek RC. 2004. Osedax: bone-eating marine worms with dwarf males. Science. 305(5684):668–671.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Zhou YD, Cheng H, Zhang DS, Wang CS. 2019. The mitochondrial genome of a slit limpet Pseudorimula sp. (Vetigastropoda: Lepetodrilidae) from hydrothermal vent on the Southwest Indian Ridge. Mitochondrial DNA B. 4(1):1189–1190.
- Zhou YD, Wang Y, Li YN, Shen CC, Liu ZS, Wang CS. 2020. First report of Osedax in the Indian Ocean indicative of trans-oceanic dispersal through the Southern Ocean. Mar Biodivers. 50(4).