Abstract
We determined the complete mitochondrial genome of Batillaria cumingi. The B. cumingi mitochondrial genome is 16,100 bp in length, comprising 13 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes. The nucleotide composition for B. cumingi is 17.5% of C, 16.88% of G, 35.3% of T, and 30.31% of A. In 13 protein-coding genes, all genes start with ATG. For the stop codon, the cox2 gene stops with TTC, the cytb, nad1, and nad2 genes stop with TAG, and the other nine genes are with TAA. Of these 37 genes identified, nine protein-coding genes and six transfer RNA genes are encoded on the heavy strand and the other genes on the light strand. The phylogenetic tree was constructed based on 13 protein-coding genes of the B. cumingi and other 19 Gastropoda species, Sepia latimanus as outgroup using the Neighbour-joining method. The tree showed that the B. cumingi is closely related to the Semisulcospira coreana in Cerithioidea. We believe that this result will be helpful for the study of population genetic and phylogenetic analysis of the family Batillariidae.
Batillaria cumingi (Crosse, 1862) belongs to the family Batillariidaeis an intertidal gastropod. It is a kind of broad-temperate benthic gastropod which distributed in Japan, Korea and northern China (Okutani and Habe Citation1983). The shell of B. cumingi is cone-shaped and strong and the shell surface has low and thin longitudinal ribs. The species is algophagous, which lives on the mud beach in the middle and upper part of the intertidal zone, where wave intensity is low (Adachi and Wada Citation1997). At present, there is no research on the mitochondrial genome of B. cumingi. In this study, it is the first report of a complete mitochondrial genome sequence of B. cumingi. The specimen of B. cumingi was collected from Haikou, Hainan province, China (110.35°E, 20.02°N) and identified by morphology and deposited in Zhejiang Ocean University. The genomic DNA extraction was utilized the salting-out method (Aljanabi and Martinez Citation1997) with the muscle, then stored at −20 °C refrigerator in the National Engineering Research Center for Marine Aquaculture, Zhejiang Ocean University (specimen Accession number: BC20181001). The genomic DNA was prepared in 400 bp paired-end libraries, and The Illumina HiSeq X Ten platform was using total genomic DNA to sequence the mitochondrial genome. All the data were available and enumerated to the Microsoft oneDrive database (https://1drv.ms/w/s!ArF1Al5lLW_VatOzZ4ygq_H6jmY?e=Z3OYmF).
The B. cumingi mitochondrial genome is 16,100 bp in length (GenBank accession number: MT323103), comprising 13 protein-coding genes, 22 transfer RNA genes, and two ribosomal RNA genes. The nucleotide composition for B. cumingi is 17.5% of C, 16.88% of G, 35.3% of T, and 30.31% of A. In 13 protein-coding genes, all genes start with ATG. For the stop codon, the cox2 gene stops with TTC, the cytb, nad1, and nad2 genes stop with TAG, and the other nine genes are with TAA. Of these 37 genes identified, nine protein-coding genes and six transfer RNA genes are encoded on the heavy strand and the other genes on the light strand. The 12S rRNA is between the tRNAThr and tRNASer, and the 16S rRNA is between the tRNAVal and tRNALeu.
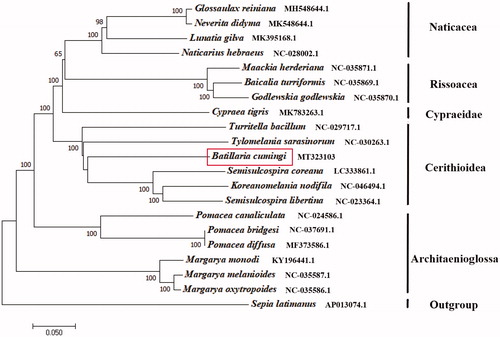
The phylogenetic tree was constructed based on 13 protein-coding genes of the B. cumingi and other 19 Gastropoda species, Sepia latimanus as outgroup using the Neighbour-joining method (Saitou and Nei Citation1987) by the program Phylip (Felsenstein Citation1989). The tree showed that the B. cumingi is closely related to the Semisulcospira coreana in Cerithioidea, similar to Cypraeidae and Architaenioglossa (). We believe that this result will be one supplement of the genome information in mitochondrial of the family Batillariidae and facilitate the study on population genetic.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in Microsoft OneDrive at https://1drv.ms/w/s!ArF1Al5lLW_VatOzZ4ygq_H6jmY?e=Z3OYmF; and in Genbank, reference number: MT323103.
Additional information
Funding
References
- Adachi N, Wada K. 1997. Distribution of Batillaria cumingi (Gastropoda, Potamididae) in Tanabe Bay, middle Japan. Nankiseibutu. 39:33–38.
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25(22):4692–4693.
- Felsenstein J. 1989. PHYLIP-phylogeny inference package (Version 3.2). Cladistics. 5:164–166.
- Okutani T, Habe T. 1983. The mollusks of Japan (sea snails). Tokyo: Gakusyukenkyusya.
- Saitou N, Nei M. 1987. The Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425.