Abstract
Cervus canadensis nannodes (Merriam, 1905) is one of the subspecies of elk distributed only in California, USA. We completed the first mitogenome of C. canadensis nannodes. Its length is 16,428 bp, which is in middle among 24 available Cervus mitogenomes. It contains 37 genes (13 protein-coding genes, 2 rRNAs, and 22 tRNAs). Phylogenetic trees show that C. c. nannodes was clustered with some subspecies of C. elaphus. Number of inter-subspecific variations between C. c. nannodes and C. e. alxaicus are relatively small in comparison to intraspecific variations of insect and fish mitogenomes and plant chloroplast genomes.
Cervus canadensis nannodes (Merriam Citation1905), a subspecies of elk, is distributed only in California, USA. This subspecies was considered as Cervus elaphus nannodes as one of the C. elaphus four subspecies (Schonewald Citation1994) based on the previous systematic descriptions (Merriam CH Citation1897; Nelson Citation1902; Merriam C Citation1905; Bailey Citation1935) as well as morphological classification studies (McCullough D Citation1969; Lowe and Gardiner Citation1989). Now, C. e. nannodes was changed to C. c. nannodes to clarify the origin of tule elk (Mizzi et al. Citation2017). Tule elk was near extinction until 1870s; however, a single breeding pair was discovered in southern San Joaquin Valley in 1874–1875 (McCullough DR et al. Citation1996) and now wild population is recovered. Because of the lack of its mitogenome despite of its available whole genome sequences (Mizzi et al. Citation2017), we completed C. c. nannodes mitogenome.
The brain tissue of C. c. nannodes collected in Namhae-gun, Gyeongsangnamdo, Korea (Voucher in Animal and Plant Quarantine Agency, Korea; APQA-R-19FC020; 36.125833 N, 128.199515E) was used for extracting DNA with DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc. Mitogenome was completed by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on Cervus elaphus alxaicus mitogenome (KU942399).
The mitogenome of C. c nannodes (GenBank accession is MT430939) is 16,428 bp, in middle size among 24 available Cervus mitogenomes (16,350 bp to 16,663 bp). It contains 37 genes (13 protein-coding genes, two rRNAs, 22 tRNAs) and GC ratio is 38.0%.
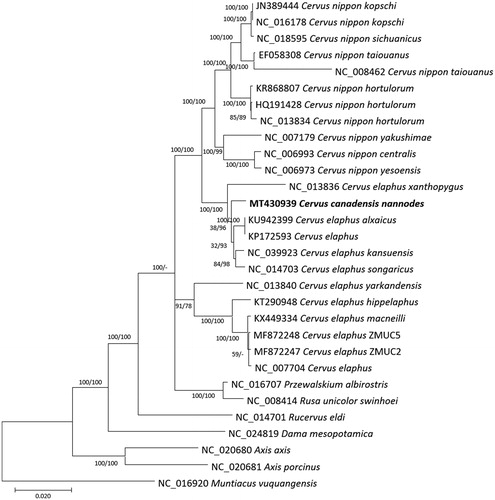
Twenty-nine complete mitogenomes of subfamily Cervinae and one outgroup species, Muntiacus vuquangensis, were used for constructing bootstrapped neighbor-joining (and maximum-likelihood trees with MEGA X (Kumar et al. Citation2018) based on complete mitogenomes alignment conducted by MAFFT 7.450 (Katoh and Standley Citation2013). Phylogenetic trees presented that C. c. nannodes was clustered together with some of the subspecies of C. elaphus with high bootstrap value, even four C. elaphus subspecies were clustered with low bootstrap value (). Inter-subspecific variations on mitogenome between C. c. nannodes and C. e. alxaicus in Cervinae display 145 SNPs and 77 INDELs, which is larger than that of inter-subspecific variations of Alces alces (subfamily Odocoileinae; 21 SNPs and 1 INDEL between KP405229 and NC_020677); however, it is smaller than intraspecific variations of mitogenomes of fishes (e.g. Rhynchocypris oxycephalus (Park, Kim, Xi Citation2019)) and insects (e.g. Chilo suppressallis (Park, Xi, et al. Citation2019), Spodoptera frugiperda (Seo et al. Citation2019), and Laodelphax striatellus (Park, Jung, et al. Citation2019)) and plant chloroplast genomes (e.g. Goodyera schlechtendaliana (Oh et al. Citation2019), Gastrodia elata (Park et al. Citation2020), Abeliophyllum distichum (Park, Kim, Xi, et al. Citation2019), and Pyrus ussuriensis (Cho et al. Citation2019)), indicating a relatively lower level of inter-subspecific variations on its mitogenomes. Moreover, phylogenetic trees displayed that the two major clades of C. elephus subspecies exists and C. nippon subspecies was separated from them ().
Figure 1. Neighbor-joining (bootstrap repeat is 10,000) and maximum-likelihood (bootstrap repeat is 1,000) phylogenetic trees of 30 complete mitogenomes: Cervus canadensis nannodes (MT430939 used in this study), Cervus elaphus alxaicus (KU942399), Cervus elaphus (NC_007704 and KP172593), Cervus elaphus kansuensis (NC_039923), Cervus elaphus songaricus (NC_014703), Cervus elaphus yarkandensis (NC_013840), Cervus elaphus hippelaphus (KT290948), Cervus elaphus macneilli (KX449334), Cervus elaphus (MF872248 and MF872247), Cervus nippon yesoensis (NC_006973), Cervus nippon centralis (NC_006993), Cervus nippon yakushimae (NC_007179), Cervus nippon hortulorum (NC_013834), Cervus nippon hortulorum (HQ191428), Cervus nippon hortulorum (KR868807), Cervus nippon taiouanus (NC_008462), Cervus nippon taiouanus (EF058308), Cervus nippon sichuanicus (NC_018595), Cervus nippon kopschi (NC_016178), Cervus nippon kopschi (JN389444), and Muntiacus vuquangensis (NC_016920) as an outgroup. Phylogenetic tree was drawn based on the maximum-likelihood tree. The numbers above branches indicate bootstrap support values of maximum-likelihood and neighbor-joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MT430939 in the NCBI GenBank.
Additional information
Funding
References
- Bailey V. 1935. A new name for the Rocky Mountain elk. Proc Biol Soc Washington. 48:187–189.
- Cho M-S, Kim Y, Kim S-C, Park J. 2019. The complete chloroplast genome of Korean Pyrus ussuriensis Maxim.(Rosaceae): providing genetic background of two types of P. ussuriensis. Mitochondrial DNA Part B. 4(2):2424–2425.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Lowe V, Gardiner A. 1989. Are the new and old world wapitis (Cervus canadensis) conspecific with red deer (Cervus elaphus)?. J Zool. 218(1):51–58.
- McCullough D. 1969. The tule elk: its history, behavior, and ecology (Vol. 88). Berkeley: University of California Publications in Zoology.
- McCullough DR, Fischer JK, Ballou JD. 1996. From bottleneck to metapopulation: recovery of the tule elk in California. Edited by D.R. McCullough. Island Press, Covelo, Calif. pp. 375–403.
- Merriam C. 1905. A new elk from California, Cervus nannodes. Proc Bioll Soc (Washington). 18:23–25.
- Merriam CH. 1897. Cervus Roosevelti: a new elk from the olympics. Washington: Biological Society of Washington.
- Mizzi JE, Lounsberry ZT, Brown CT, Sacks BN. 2017. Draft genome of tule elk Cervus canadensis nannodes. F1000Res. 6:1691.
- Nelson EW. 1902. A new species of elk from Arizona. Vol. 16. Order of the Trustees. New York : Published by order of the Trustees, American Museum of Natural History https://digitallibrary.amnh.org/handle/2246/748?show=full
- Oh S-H, Suh HJ, Park J, Kim Y, Kim S. 2019. The complete chloroplast genome sequence of Goodyera schlechtendaliana in Korea (Orchidaceae). Mitochondrial DNA Part B. 4(2):2692–2693.
- Park J, Jung JK, Ho Koh Y, Park J, Seo BY. 2019. The complete mitochondrial genome of Laodelphax striatellus (Fallén, 1826) (Hemiptera: Delphacidae) collected in a mid-Western part of Korean peninsula. Mitochondrial DNA Part B. 4(2):2229–2230.
- Park J, Kim Y, Xi H. 2019. The complete mitochondrial genome sequence of Chinese minnow in Korea, Rhynchocypris oxycephalus (Sauvage and Dabry de Thiersant, 1874). Mitochondrial DNA Part B. 4(1):662–663.
- Park J, Kim Y, Xi H, Jang T, Park J-H. 2019. The complete chloroplast genome of Abeliophyllum distichum Nakai (Oleaceae), cultivar Ok Hwang 1ho: insights of cultivar specific variations of A. distichum. Mitochondrial DNA Part B. 4(1):1640–1642.
- Park J, Suh Y, Kim S. 2020. A complete chloroplast genome sequence of Gastrodia elata (Orchidaceae) represents high sequence variation in the species. Mitochondrial DNA Part B. 5(1):517–519.
- Park J, Xi H, Kwon W, Park C-G, Lee W. 2019. The complete mitochondrial genome sequence of Korean Chilo suppressalis (Walker, 1863) (Lepidoptera: Crambidae). Mitochondrial DNA Part B. 4(1):850–851.
- Schonewald C. 1994. Cervus canadensis and C. elaphus: North American subspecies and evaluation of clinal extremes. Acta Theriol. 39:431–431.
- Seo BY, Lee G-S, Park J, Xi H, Lee H, Lee J, Park J, Lee W. 2019. The complete mitochondrial genome of the fall armyworm, Spodoptera frugiperda Smith, 1797 (Lepidoptera; Noctuidae), firstly collected in Korea. Mitochondrial DNA Part B. 4(2):3918–3920.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
