Abstract
In this study, we first sequenced and characterized the complete mitochondrial genome of Onychostoma leptura. The complete mitochondrial genome was 16,601 bp in length and included 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), two ribosomal RNA genes (12S rRNA and 16S rRNA), and one non-coding control region (D-loop). The overall nucleotide composition was A: 31.31%, T: 23.89%, G:16.16%, and C: 28.64%, and the A + T content (55.20%) was much higher than G + C content (44.80%). The phylogenetic tree indicated that nine species belong to Onychostoma in the present study were classified into two clades. These results can provide basic genetic information for population genetic research of O. leptura and phylogenetic analysis of Onychostoma.
Onychostoma leptura (genus: Onychostoma, subfamily: Barbinae, family: Cyprinidae), an endemic fish in East Asia, is mainly distributed in Laos, Vietnam, and China (Kottelat Citation2001). It has a slender and laterally compressed body, a thin caudal peduncle, and a dark stripe along the lateral line of the body. The edges of anal, dorsal, and caudal fins of O. leptura are light red (Yue Citation2000). It feeds on scraping algae attached to rocks and inhabits streams with turbulent and clear water (Kottelat Citation2001). It has a certain population and is the main economic fish in some places, such as Hainan Island of China (Zhou et al. Citation2016). Here, we first sequenced the complete mitochondrial genome of O. leptura, which will provide basic genetic information for population genetic research of O. leptura and phylogenetic analysis of Onychostoma.
Specimen of O. leptura was collected from Shibizhen (N19°09′46″ E110°18′30″) section of the Wanquan River in Hainan Island in 2018. The collected specimen was deposited in 95% alcohol and stored at −20 °C in Specimen Room of School of Life Sciences, Jianghan University (Specimen code: O. leptura 20180413001). The total genomic DNA was extracted from dorsal muscle by using the animal tissue DNA kit. The complete mitochondrial genome was sequenced using the Illumina high-throughput sequencing technology and de novo assembled using NOVOPlasty ver 2.6 (Dierckxsens et al. Citation2017). Protein-coding genes and rRNA genes were predicted using web server DOGMA (Wyman et al. Citation2004), and tRNA genes were created using the program ARWEN (Laslett and Canback Citation2008).
The complete mitochondrial genome of O. leptura was 16,601 bp in length (NCBI GenBank accession number: MT449680). The overall nucleotide composition was A: 31.31%, T: 23.89%, G:16.16%, and C: 28.64%, and the A + T content (55.20%) was much higher than G + C content (44.80%). The complete mitochondrial genome included 13 protein-coding genes (ND1, ND2, ND3, ND4, ND4L, ND5, ND6, COI, COII, COIII, ATP6, ATP8, Cyt b), 22 transfer RNA genes (tRNA), two ribosomal RNA genes (12S rRNA and 16S rRNA), and one non-coding control region (D-loop). All the genes were encoded on the heavy strand, except for one protein-coding gene (ND6) and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu and tRNAPro) were encoded on the light strand.
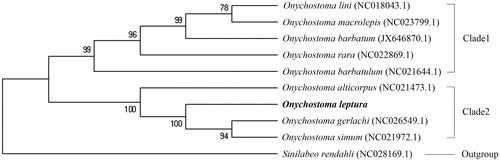
A phylogenetic tree was constructed using Neighbour-Joining method using MEGA7 (Kumar et al. Citation2016) based on the whole mitogenome sequences of nine species belong to Onychostoma and Sinilabeo rendahli that was used as the outgroup (). The complete mitochondrial genome of other species used for the construction of phylogenetic tree were downloaded from NCBI GenBank. The phylogenetic tree indicated that nine species belong to Onychostoma in the present study were classified into two clades: O. lini, O. macrolepis, O. barbatum, O. rara, and O. barbatulum in clade1, O. alticorpus, O. leptura, O. gerlachi, and O. simum in clade2.
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT449680.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Kottelat M. 2001. Fishes of Laos. Colombo: WHT Publications Ltd; p. 198.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20(17):3252–3255.
- Yue P, editor. 2000. Fauna Sinica, Osteichthyes, CypriniformesII. Beijing: Science Press (in Chinese).
- Zhou TQ, Lin HD, Hsu KC, Kuo PH, Wang WK, Tang WQ, Liu D, Yang JQ. 2016. Spatial genetic structure of the cyprinid fish Onychostoma lepturum on Hainan Island. Mitochondrial DNA A. 28(6):1–8.