Abstract
The complete mitochondrial genome of Chinapotamon maolanense was obtained for the first time. The complete mitochondrial genome of C. maolanense is 17,130 bp in length, including 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 1 control region. In addition, the mitogenome has 18 noncoding regions ranging from 1 to 1553 bp in length.
The species Chinapotamon maolanense (Dai Citation1999) (Crustacea: Malacostraca:Decapoda: Brachyura: Potamidae: Chinapotamon) is distributed in the karst landform area of Guizhou Province, China (Jie-Xin et al.). The complete mitochondrial genome of C. maolanense was obtained for the first time.
An adult specimen of C. maolanense was collected from Panzhai Village, Dawn township aquatic animals, Libo County, Qiandongnan Miao and Dong Autonomous Prefectu, Guizhou Province, China in 2017 (N25.234444°, E 108. 029527°). The sample has been deposited in the Laboratory Specimen Library of Freshwater Crustacean Decapoda & Paragonimus, School of Basic Medical Sciences, Nanchang University, Nanchang, Jiangxi, PR China & National Parasite Germplasm Resources Specimen Library of China with a catalogue number of NCUMCP1960. The sample was stored in 95% ethanol prior to extraction at room temperature before sequence analyses. Genomic DNA extraction, sequencing, gene annotation, and phylogenetic analyses were performed according to the method described by Plazzi et al. (Citation2013). The Bayesian Inference (BI) method was performed using MrBayes vers. 3.2 (Ronquist et al. Citation2012), with best model GTR + I + G selected using jModelTest vers. 2.1.7. The maximum-likelihood (ML) method was performed using MEGA 6 (Tamura et al. Citation2013).
Chinapotamon maolanense (GenBank accession no. MT134100) has a full-length mitochondrial genome of 17,130 bp and shares the same 37 genes (13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes) and 1 control region (CR) as the typical metazoan mitochondrial genome (Yan et al.), with a high A + T bias (71.7%). The CR of C. maolanense is located in a typical crustacean position (between 12SrRNA and tRNAIle), it is 1553 bp in length, and has a higher A + T bias (76.5%) than does the mitochondrial genome. Among these genes, 23 are located in the H-strand, and 14 are located in the L-strand.
The mitochondrial genome of C. maolanense has 18 noncoding regions and is between 1–1553 bp in length, with the longest noncoding region between 12S rRNA and tRNAIle which is considered to be the CR; there are nine gene overlap areas, with a total length of 25 bp, and those areas are between 1 and 7 bp in length, with the longest gene overlap area between ND4 and ND4L.
The mitochondrial genome of C. maolanense contains 13 protein-coding genes. Similar to other Brachyura mitochondrial genomes, 9 protein-coding genes are located in the H chain (COX1, COX2, COX3, ATP6, ATP8, ND2, ND3, ND6, and CYTB), and the remaining four are located in the L chain (ND1, ND4, ND4L and ND5). The initiator codon of protein-coding genes are ATG (COX1, COX2, ATP8, COX3, ND4, ND4L, Cyt b, ND1 and ND2), and ATA (ATP6, ND3, ND5 and ND6); the termination codon are TAA (COX1, COX2, ATP6, COX3, ND4, ND4L, ND6, ND1 and ND6), and TAG (ATP8), and incomplete termination codon T (ND3, ND5 and Cyt b). The A + T bias of the protein-coding gene is 69.0%.
The mitochondrial genome of C. maolanense possesses two rRNA genes, 16S rRNA and 12S rRNA, which are located in the same L chain as other Brachyura mitochondrial genomes. The length of the 16S rRNA and 12S rRNA genes is 1318 bp and 824 bp, respectively; 16S rRNA is between tRNALeu(CUN) and tRNAVal, while 12S rRNA is between tRNAGln and the CR.
Chinapotamon maolanense has 22 tRNAs in common, and most tRNAs have a typical clover structure, with the exception of tRNASer(AGN) which lacks the dihydrouracil (DHU) arm (Ohtsuki et al. Citation2002). The 22 tRNA genes are between 61 (tRNAArg) and 74 bp (tRNAVal) in length. Additionally, there are certain base mismatches, including 36G–T mismatches, 3A–C mismatches, 1A–G mismatches, 1T–C mismatches, 1A–A mismatches, and 2T–T mismatches.
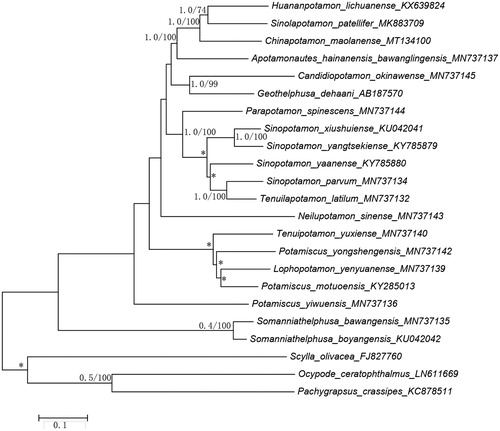
The phylogenetic position of C.maolanense in mitogenome relative to other freshwater crabs mitogenomes is determined by applying the BI and ML methods on 13 PCGs (). Our results indicate that as one of the freshwater crabs, C. maolanense is the sister group with Huananpotamon lichuanense and Sinolapotamon patellifer. This result is consistent with morphological classification and other molecular analyses (Dai Citation1999).
Figure 1. Bayesian Inference (BI) phylogenetic tree of Chinapotamon maolanense and other related freshwater crabs based on 13 PCGs in mitogenomes. Numbers on internodes are BI bootstrap proportions and the ML posterior proportions. The same of phylogenetic trees between ML and BI are indicated by bold branches. The differences of freshwater crabs between the ML and BI trees are indicated by ‘*’.The scale bars represent genetic distance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT134100.
Additional information
Funding
References
- Dai AY. 1999. Fauna sinica, Arthropoda, Crustacea, Malacostraca, Decapoda, Parathelphusidae Potamidae. Beijing, China: Science Press.
- Jie-Xin Z, Jun B, Xian-Min Z. A new species of karst-dwelling freshwater crab of the genus Chinapotamon Dai & Naiyanetr, 1994 (Crustacea: Decapoda: Brachyura: Potamidae), from Guizhou, southwest China.
- Ohtsuki T, Kawai G, Watanabe K. 2002. The minimal tRNA: unique structure of Ascaris suum mitochondrial tRNASerUCU having a short T arm and lacking the entire D arm. Febs Lett. 514(1):37–43.
- Plazzi F, Ribani A, Passamonti M. 2013. The complete mitochondrial genome of Solemya velum (Mollusca: Bivalvia) and its relationships with Conchifera. BMC Genom. 14(1):409.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Yan W, Zeng QW, Zou JX, Zhu CC, Zhou XM. The complete mitochondrial genome of freshwater crab Sinopotamon xiushuiense (Decapoda: Brachyura: Potamoidea).
