Abstract
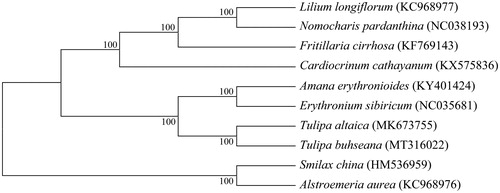
The complete chloroplast genome of Tulipa buhseana was sequenced and reported here. The circular genome of T. buhseana is 152,062 bp in length and contains 133 functional genes consisting of 87 coding sequences, 38 tRNA genes, and 8 rRNA genes. With 1 species from Smilacaceae and 1 species from Alstroemeriaceae as outgroup, phylogenetic relationships of 8 Liliaceae species based on their chloroplast genomes indicated that T. buhseana is closest to T. altaica.
Tulipa L. belongs to the Liliaceae family and includes approximately 139 species in the world (Zhou et al. Citation2019), which is one of the world’s most well-known, beloved, and economically important flowering plants (Li et al. Citation2017). Tulipa L. is widely distributed in Central Asia, the Mediterranean, Europe and North Africa (Xing et al. Citation2017). Here, we sequenced the complete chloroplast genome of Tulipa buhseana to provide more fundamental data to the development of the DNA barcoding system.
In this study, T. buhseana were collected from Altay Region, Buerjing County, Xinjiang Province, China (48°41′48″N, 87°02′03″E) in September 2018. The specimen was kept in the Key Laboratory of Landscape Plants Lab, College of Agriculture and Animal Husbandry, Qinghai University, Xining, China (accession number: JXT-2018-YJX016). Genomic DNA was extracted using leaves from the same plant. Genomic sequencing was performed on the Illumina HiSeq Platform (Illumina, San Diego, CA) with a read length of 150 bp. The software SPAdes v.3.14.0 (Bankevich, et al. Citation2012) was employed to assemble the chloroplast genome. Then, Prodigal v2.6.3 (https://www.github.com/hyattpd/Prodigal), Hmmer v3.1b2 (http://www.hmmer.org/) and Aragorn v1.2.38 (http://130.235.244.92/ARAGORN/) were respectively used to annotate the coding sequences (CDs), transfer RNA (tRNA) genes, and ribosomal RNA (rRNA) genes.
A circularized DNA assembly was 152,062 bp in length. It was harvested and deposited in the GenBank with accession number of MT316022. Totally, 133 functional genes are recognized in this chloroplast genome: 87 CDs, 38 tRNA genes, and 8 rRNA genes. The rRNA genes, tRNA genes, and protein-coding genes account for 6.02%, 28.57%, and 65.41% of all annotated genes, respectively. The GC content of the complete chloroplast genome was 36.61%.
Based on chloroplast genomes assembled here and downloaded from GenBank, phylogenetic relationships of 8 Liliaceae species were resolved by means of neighbor-joining with 1 species from Smilacaceae and 1 species from Alstroemeriaceae as outgroup (). After aligned using MAFFT (Katoh and Standley Citation2013), the neighbor-joining tree was built using MEGA7 (Kumar et al. Citation2016) with bootstrap set to 1000. In the phylogenetic tree, T. buhseana and T. altaica went to an independent clade with a 100% node support rate.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in Genbank at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT316022.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li P, Lu RS, Xu WQ, Ohi-Toma T, Cai MQ, Qiu YX, Cameron KM, Fu CX. 2017. Comparative genomics and phylogenomics of East Asian Tulips (Amana, Liliaceae). Front Plant Sci. 8:1–12.
- Xing GM, Qu LW, Zhang YQ, Xue L, Su JW, Lei JJ. 2017. Collection and evaluation of wild tulip (Tulipa spp.) resources in China. Genet Resour Crop Evol. 64(4):641–652.
- Zhou JT, Yin PP, Chen Y, Zhao YP. 2019. The complete chloroplast genome of Tulipa altaica (Liliaceae), a wild relative of tulip. Mitochondrial DNA Part B. 4(1):2017–2018.