Abstract
Fusarium oxysporum f. sp. KGSJ26F3 is a plant pathogenic filamentous fungus isolated from wilted potato in northwest China where potato is the most important crop. We carried out a complete mitochondrial genome of F. oxysporum. The results showed the circular molecule is 46,664 bp, and the base composition of the mitogenome is as follows: A (34.3%), T (33.6%), C (14.6%), and G (17.5%). The mitogenome contains 18 protein-coding genes, two ribosomal RNA (rRNA), and 26 transfer RNA (tRNA) genes. The gene order is identical to that of the other Fusarium mitogenomes. The taxonomic status of the F. oxysporum mitogenome exhibits a closest relationship with F. oxysporum. However, it varied in the structure of mitochondrial genome.
Fusarium oxysporum is a soil borne fungal pathogen which can infect a variety of crops and cause serious crop yield reduction (Fravel et al. Citation2003). There are more than 120 species of F. oxysporum (Armstrong Citation1981). With the advancement of scientific research and time, over 63 F. oxysporum have been sequenced and compared their mitotic genome. The mitochondrial genome of F. oxysporum is diverse and has many different characteristics from that of other fungi (Brankovics et al. Citation2017; Chen et al. Citation2019; Park, Kwon, Huang, et al. Citation2019; Park, Kwon, Zhu, et al. Citation2019). Fusarium oxysporum strain KGSJ26F3(GenBank accession number MK764912) was isolated from wilted potato in Gansu, northwest China (35°26.346′N, 104°50.991′E) and identified based on translation elongation factor 1a gene and internal transcribed spacer (White et al. Citation1990; Samson et al. Citation2004). The voucher specimen (No. KGSJ26F3) was deposited at the Gansu Provincial Key Lab of Arid land Crop Science, Lanzhou, Gansu, China. The total genomic DNA mycelia obtained was extracted using Fungal DNA Kit D3390-02 (Omega Bio-Tek, Norcross, GA) according to the manufacturer’s instructions and was stored in the sequencing company (Xuan Chen Biological Technology Co., Ltd.Shaanxi, China). Purified DNA was used to construct the sequencing libraries following the instructions of NEBNext®Ultra™II DNA Library Prep Kit (NEB, Beijing, China). Whole-genome sequencing was performed using the Illumina novaseq6000 Platform (Illumina, San Diego, CA). Multiple steps were used for quality control and de novo assembly of the mitogenome (Bi Citation2017). Adapters and low-quality reads were removed using the NGS QC Toolkit (Patel and Jain Citation2012).
The obtained clean reads were screened out by Bowtie 2 (Langmead and Salzberg Citation2012) with the help of a homologous reference sequence (Fusarium commune: LT906348) and then assembled as implemented by NOVOPlastyv3.8.3 (Dierckxsens et al. Citation2017). Genome annotation is mainly carried out by comparing the mitochondrial genome (see the phylogenetic tree for details) of the same genus and relative species of GenBank. Geneious R11 software (https://www.genetic.com) was used to make mitochondrial genome map. The length of F. oxysporum KGSJ26F3 mitogenome is 46,314 bp, and its size, structure, and gene content are similar to those of Fusarium. This mitogenome was submitted to GenBank database under accession no. MT269799. The circular mitogenome contains 18 protein-coding genes, two ribosomal RNA (rRNA), and 26 transfer RNA (tRNA) genes (). The base composition of the genome is as follows: A (34.3%), T (33.6%), C (14.6%), and G (17.5%).
Figure 1. Phylogenetic relationships among 13 Fusarium mt genomes. This tree was drawn with Potato Fusarium oxysporum as an out-group. The length of branch represents the divergence distance.
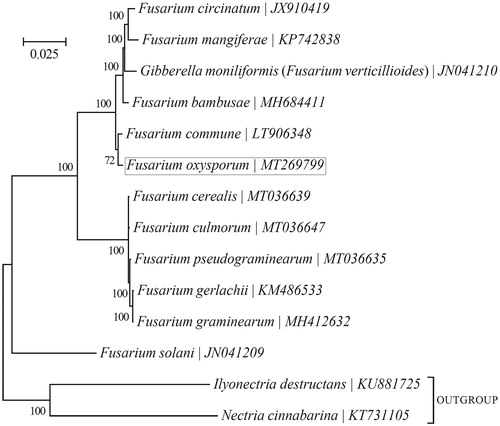
To validate the phylogenetic position of F. oxysporum, the genome-wide alignment of Fusarium mitogenomes was constructed using Geneious R11 software (https://www.genetic.com). Genes with good comparison effects were selected and then connected into a single super gene. It was exported to topali v2.5 software (Milne et al. Citation2009) to build phylogenetic tree ().
As shown in , Fusarium circinatum (JX910419), Fusarium mangiferae (KP742838), Gibberella moniliformis (JN041210), Fusarium bambusae (MH684411), and Fusarium commune (LT906348) are determined as sisters of F. oxysporum with strong support. High bootstrap and posterior probability values show that presented relations are stable. The mitochondrial genome of F. oxysporum will contribute to the understanding of phylogeny.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
We confirm that the data supporting the findings of this study are available within the article and its supplementary materials. The data that support the findings of this study are openly available in NCBI GenBank database at https://www.ncbi.nlm.nih.gov/nuccore/ with the accession number is MT269799, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Additional information
Funding
References
- Armstrong G. 1981. Formae speciales and races of Fusarium oxysporum causing wilt disease. Fusarium: disease, biology, and taxonomy. University Park (PA): Pennsylvania State University Press; p. 391–399.
- Bi GQ. 2017. The complete mitochondrial genome of northern grasshopper mouse (Onychomys leucogaster). Mitochondrial DNA B. 2(2):393–394.
- Brankovics B, van Dam P, Rep M, de Hoog GS, J van der Lee TA, Waalwijk C, van Diepeningen AD. 2017. Mitochondrial genomes reveal recombination in the presumed asexual Fusarium oxysporum species complex. BMC Genomics. 18(1):735.
- Chen C, Fu R, Wang J, Hu R, Li X, Luo X, Chen X, Lu D. 2019. Characterization and phylogenetic analysis of the completemitochondrial genome of Aspergillus sp. (Eurotiales: Eurotiomycetidae). Mitochondrial DNA B. 4(1):752–753.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fravel D, Olivain C, Alabouvette C. 2003. Fusarium oxysporum and its biocontrol. New Phytol. 157(3):493–502.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25(1):126–127.
- Park J, Kwon W, Huang X, Mageswari A, Heo I-B, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of a xerophilic fungus Aspergillus pseudoglaucus. Mitochondrial DNA B. 4(2):2422–2423.
- Park J, Kwon W, Zhu B, Mageswari A, Heo I-B, Kim J-H, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of an aflatoxin B and G producing fungus Aspergillus parasiticus. Mitochondrial DNA B. 4(1):947–948.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7(2):e30619.
- Samson RA, Hoekstra ES, Frisvad JC. 2004. Introduction to food-and airborne fungi. In: Tjamos EC, Papavizas GC, Cook RJ, editors. Centraal bureau voor Schimmelcultures (CBS). 7th ed. Boston (MA).
- White TJ, Sninsky JJ, Gelfand DH, Innis MA. 1990. Page 315 in: PCR protocols: a guide to methods and applications. San Diego: Academic Press.
