Abstract
The complete chloroplast genome sequence of Semiaquilegia guangxiensis was assembled and the phylogenetic relationship with other species in Trib. Isopyreae was inferred in this study. The chloroplast genome is 164,047 bp in length. A typical quadripartite structure was detected, including two inverted repeats (IRs) of 29,581 bp, which are separated by a large single-copy (LSC) and a small single-copy (SSC) of 87,490 bp and 17,395 bp, respectively. Moreover, The genome comprises 132 genes, including 88 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content of the chloroplast genome is 38.9%. The phylogenetic analysis indicated that S. guangxiensis is most closely related to its congener S. adoxoides, and Semiaquilegia is most closely related to Aquilegia in Ranunculaceae.
Semiaquilegia Makino was assumed to consist of a sole species in the family Ranunculaceae till to the recent past, namely S. adoxoides (DC.) Makino – a perennial medicinal herb with tuberous root. It is distributed in Asian subtropic and temperate regions in China, Korea, and Japan, growing in various habitats of fields, roadsides, valleys, slopes, etc. (Fu and Orbélia Citation2001). In recent years, three new species of Semiaquilegia, namely S. guangxiensis Yan Liu & Y.S. Huang, S. quelpaertensis D.C. Son & K. Lee, and S. danxiashanensis L. Wu, J.J. Zhou, Q. Zhang & W.S. Deng have been described (Huang et al. Citation2017; Son et al. Citation2017; Zhou et al. Citation2019). Among them, S. guangxiensis is endemic to broad-leaved forests of limestone hills in Guangxi, China; S. quelpaertensis grows in submontane broad-leaved forests in Korea; and S. danxiashanensis is restricted to Danxia landform in Guangdong, China. However, the complete chloroplast genomes have not been reported in this genus yet except for S. adoxoides (Zhai et al. Citation2019). Here, we report the complete chloroplast genome of S. guangxiensis, and the inferred phylogenetic relationships of this species as well as the genus Semiaquilegia to other taxa in Trib. Isopyreae.
Total DNA was extracted from the silica-dried leaves of S. guangxiensis, which was collected from Yongfu county, Guangxi, China (24°56′10″N, 110°6′46″E). The voucher specimen was deposited at the Herbarium of Guangxi Institute of Botany (IBK00421269). The genomic paired ends (PE150) sequencing was performed on NovaSeq 6000 (in Novogene corp., Tianjin, China). Approximately 2.4 Gb of clean data were obtained. De novo assembly were employed with SPAdes 3.11.0 (Bankevich et al. Citation2012). Annotation was performed using PGA (Qu et al. Citation2019) with reference to S. adoxoides (MK569498).
The chloroplast genome of S. guangxiensis (GenBank accession number: MT410682) has a total length of 164,047 bp with an overall GC content of 38.9%. The cp genome contains one LSC region of 87,490 bp and one SSC region of 17,395 bp, which are separated by two IR regions of 29,581 bp, respectively. A total of 132 genes are encoded for the chloroplast genome, which include 88 protein-coding genes, 8 rRNA genes, and 36 tRNA genes. In these genes, six tRNA genes (trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC) and nine protein-coding genes (atpF, ndhA, ndhB, petB, petD, rps16, rpl16, rpl2, and rpoC1) contain one intron, each of the two gene (clpP, ycf3) has two introns, and rps12 gene has trans-splicing.
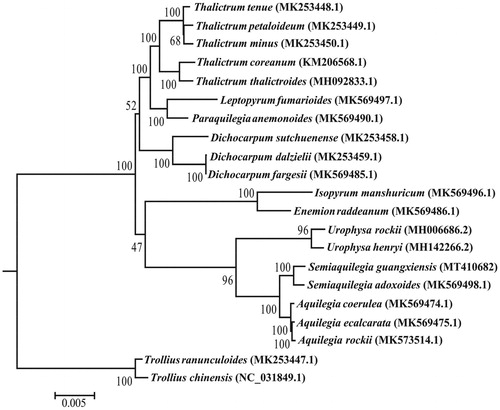
To understand the phylogenetic position of S. guangxiensis as well as the genus Semiaquilegia within Trib. Isopyreae in the family Ranunculaceae, a maximum likelihood tree was constructed by RAxML (Stamatakis Citation2014) based on the concatenated data of 74 protein-coding genes from the chloroplast genome sequences of 21 species with Trollius chinensis and T. ranunculoides selected as outgroups according to the larger-scale phylogenetic studies in Ranunculaceae (Zhai et al. Citation2019). The phylogenetic tree shows that S. guangxiensis is most closely related to S. adoxoides (BS = 100%), and Semiaquilegia is most closely related to Aquilegia in Ranunculaceae (BS = 100%) ().
Disclosure statement
No potential conflict of interest was declared by the author(s).
Data availability statement
The complete chloroplast genome sequence of S. guangxiensis has been submitted to the GenBank (https://www.ncbi.nlm.nih.gov/genbank/), and the accession number is MT410682. This sequence will be released immediately after process by the NCBI staff. Then, the data that support the findings of this study is openly available in GenBank.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Fu DZ, Orbélia RR. 2001. Semiaquilegia Makino. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, Vol. 6. Beijing: Science Press, and St. Louis: Missouri Botanical Garden Press. pp. 281–282.
- Huang YS, Guo J, Zhang Q, Lu ZC, Liu Y. 2017. Semiaquilegia guangxiensis (Ranunculaceae), a new species from the limestone areas of Guangxi, China, based on morphological and molecular evidence. Phytotaxa. 292(2):180–188.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Son DC, Jeong KS, Lee KH, Kim H, Chang KS. 2017. Semiaquilegia quelpaertensis (Ranunculaceae), a new species from the Republic of Korea. PK. 89(1):107–113.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhai W, Duan XS, Zhang R, Guo C, Li L, Xu GX, Shan HY, Kong HZ, Ren Y. 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135(2019):12–21.
- Zhou JJ, Huang ZP, Li JH, Hodges S, Deng WS, Wu L, Zhang Q. 2019. Semiaquilegia danxiashanensis (Ranunculaceae), a new species from Danxia Shan in Guangdong, southern China. Phytotaxa. 405(1):1–014.