Abstract
The Tangut monkshood (Aconitum tanguticum) is a perennial herb with high medicinal values. Here, its chloroplast genome was assembled from Illumina sequencing reads. The circular genome is 157,114 bp long with an A + T-biased nucleotide composition, and comprises a pair of inverted repeat (IR) regions (26,255 bp), separated by a large single-copy (LSC) region (87,559 bp) and a small single-copy (SSC) region (17,045 bp). It encodes a total of 112 gene species, with 19 of them being completely or partially duplicated and 18 of them harboring one or two introns. Phylogenetic analysis recovered two major clades of the genus Aconitum.
Aconitum tanguticum, commonly known as Tangut monkshood, is a perennial herb within the family Ranunculaceae (Ranunculales), and is mainly distributed in Gansu, Qinghai, Shaanxi, Sichuan, Tibet and Yunnan Provinces of China with an elevation of 3200–4800 m (Li and Yuichi Citation2001). This herb has long been used in traditional Tibetan medicine for treating gastritis, hepatitis, influenza, nephritis, pneumonia and other diseases (Nanjing University of Traditional Chinese Medicine Citation2005). To date, most studies of A. tanguticum have been focused upon its phytochemistry (e.g. Xu et al. Citation2013; Li et al. Citation2014, Citation2015). Little is known about its genomics. In this study, its complete chloroplast genome was assembled from high-throughput Illumina sequencing reads. The annotated sequence was deposited into GenBank under the accession number MT430949.
Fresh leaves of a single individual were collected from Laji Mountain (101°47′11″E, 36°02′14″N) with the voucher specimen deposited in Qinghai University (accession number: LQE-2019-066), and were used for genomic DNA extraction with the DNeasy Plant Mini Kit (Qiagen, CA, USA). The high-throughput DNA sequencing was performed on the Illumina HiSeq X Ten Sequencing System (Illumina, CA, USA), and yielded a total of 94.97 M of 150-bp raw paired reads. The chloroplast genome was assembled using MITObim v1.9 (Hahn et al. Citation2013) with that of Aconitum carmichaelii (Yang et al. Citation2018) as the initial reference. Genomic annotation was done in Geneious Prime 2020 (Biomatters Ltd., Auckland, New Zealand) by aligning with those of its congeners, e.g. Aconitum delavayi (MG678802) (Meng et al. Citation2018), Aconitum chiisanense (KT820665) (Lim et al. Citation2017) and Aconitum reclinatum (MF186593) (Kong et al. Citation2018).
The chloroplast genome of A. tanguticum is 157,114 bp in size, and comprises a pair of inverted repeat (IR) regions (26,255 bp), separated by a large single-copy (LSC) region (87,559 bp) and a small single-copy (SSC) region (17,045 bp). The nucleotide composition is asymmetric with an overall A + T content of 62.0% (‘light strand’). In all, 112 gene species were annotated, including 78 protein-coding (PCG), 30 tRNA and four rRNA gene species. Nineteen gene species are completely or partially duplicated, including eight PCGs (ndhB, rpl2, rpl23, rps7, rps12, ycf1, ycf2 & ycf15), seven tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG & trnV-GAC) and all four rRNAs (rrn4.5, rrn5, rrn16 & rrn23). Besides, one or two introns are present in 18 gene species (i.e., atpF, clpP, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, rps16, ycf3, trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC).
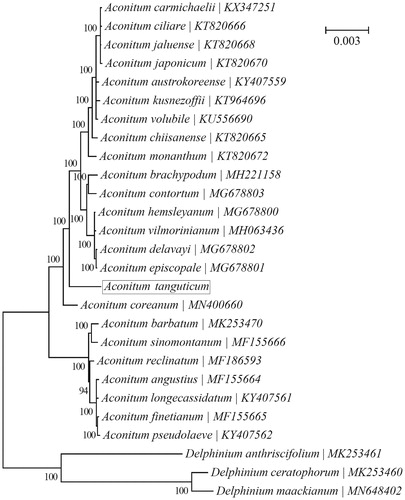
A Bayesian phylogeny was reconstructed using chloroplast PCGs for a panel of 24 species within the genus Aconitum with the program MrBayes v3.1.1 (Huelsenbeck and Ronquist Citation2001; Ronquist and Huelsenbeck Citation2003) (). ‘GTR + G+I’ was selected as the best-fit nucleotide substitution model by TOPALi v2.5 (Milne et al. Citation2009). Three species within the genus Delphinium, i.e. D. maackianum (MN648402) (He et al. Citation2019), D. anthriscifolium (MK253461) and D. ceratophorum (MK253460) (Park et al. Citation2020), were included as outgroup taxa. All 24 species were clustered into two major clades, with one clade consisting of seven species (A. angustius, A. barbatum, A. finetianum, A. longecassidatum, A. pseudolaeve, A. reclinatum & A. sinomontanum) and the other consisting of the remaining 17 species. A. tanguticum was placed within the latter larger clade.
Figure 1. Phylogenetic relationships of 24 species within the genus Aconitum based on the Bayesian analysis of the concatenated coding sequences of chloroplast PCGs. The best-fit nucleotide substitution model is ‘GTR + G+I’. Three contribal species within the genus Delphinium were included as outgroup taxa.
Disclosure statement
No potential conflict of interest was reported by the author(s)
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT430949.
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucl Acids Res. 41(13):e129–e129.
- He J, Yao M, Lyu R-D, Lin L-L, Liu H-J, Pei L-Y, Yan S-X, Xie L, Cheng J. 2019. Structural variation of the complete chloroplast genome and plastid phylogenomics of the genus Asteropyrum (Ranunculaceae). Sci Rep. 9(1):15285.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Kong H, Liu W, Yao G, Gong W. 2018. Characterization of the whole chloroplast genome of a rare and endangered species Aconitum reclinatum (Ranunculaceae) in the United States. Conserv Genet Resour. 10(2):165–168.
- Li Y-R, Liu T, Yan R-Y, Hui L-Q, Lin L-M, Cao C-Y, Guo S-S, Yang L-X, Feng W-H, Li C, et al. 2015. Three new phenolic glycosides from the whole plant of Aconitum tanguticum (Maxim.) Stapf. Phytochem Lett. 11:311–315.
- Li Y-R, Xu L, Li C, Wang Z-M, Yang L-X. 2014. Two new compounds from Aconitum tanguticum. J Asian Nat Prod Res. 16(7):730–734.
- Li L, Yuichi K. 2001. Aconitum Linnaeus. Flora China. 6:149–222.
- Lim CE, Kim G-B, Baek S, Han S-M, Yu H-J, Mun J-H. 2017. The complete chloroplast genome of Aconitum chiisanense Nakai (Ranunculaceae). Mitochondrial DNA A DNA Mapp Seq Anal. 28(1):75–76.
- Meng J, Li X, Li H, Yang J, Wang H, He J. 2018. Comparative analysis of the complete chloroplast genomes of four Aconitum medicinal species. Molecules. 23(5):1015.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25(1):126–127.
- Nanjing University of Traditional Chinese Medicine. 2005. Dictionary of Chinese medicine. Shanghai: Shanghai Science and Technology Press.
- Park S, An B, Park S. 2020. Recurrent gene duplication in the angiosperm tribe Delphinieae (Ranunculaceae) inferred from intracellular gene transfer events and heteroplasmic mutations in the plastid matK gene. Sci Rep. 10(1):2720.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Xu L, Zhang X, Lin L-M, Li C, Wang Z-M, Luo Y-M. 2013. Two new flavonol glycosides from the Tibetan medicinal plant Aconitum tanguticum. J Asian Nat Prod Res. 15(7):737–742.
- Yang J, Zeng X, Guo S. 2018. Characterization of the complete chloroplast genome of the perennial herb Aconitum carmichaelii (Ranunculales: Ranunculaceae). Conserv Genet Resour. 10(4):605–608.
