Abstract
Akebia trifoliata, commonly known as ‘Bayuezha’ in China, has been widely used as traditional Chinese medicinal herbs with a long history. In the present study, the complete chloroplast genome of A. trifoliata was sequenced using Illumina high-throughput sequencing approach. The length of the complete chloroplast genome is 157,952 bp with 38.7% GC content. It contains 131 genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis indicated that A. trifoliata was closely related to another Lardizabalaceae species, Akebia quinata, which further confirms traditional species classification.
Akebia trifoliata, a fast-growing climbing vine, belongs to Lardizabalaceae family, which is widely distributed in eastern Asian countries, including China and Japan (Li, Xiaohong et al. Citation2010). As traditional Chinese medicinal herbs, A. trifoliata has been used to promote diuresis, and activate blood circulation (Jiang et al. Citation2012). In China, A. trifoliata has also been domesticated as a fruit crop, and cultivated in Hunan, Jiangxi and Shanxi provinces (Li, Chen et al. Citation2010). Compared to those of pear, strawberry and apple, the fruits of A. trifoliata are rich in minerals and various vitamins (Li, Xiaohong et al. Citation2010; Zou et al. Citation2019). Previous studies of A. trifoliata mainly concentrated on the cultivation and phytochemical studies (Wang et al. Citation2015; Xu et al. Citation2016). Whereas, its phylogenetic analysis is lacking.
In this study, using Illumina high-throughput sequencing approach, the complete chloroplast (cp) genome of A. trifoliata was sequenced and annotated, which could facilitate the elucidation of phylogenetic evolutionary aspects in the cp genome-wide level in A. trifoliata and its closely related species.
The fresh green leaves of A. trifoliata cv. No.2 Zibao were collected from Fruit Research Institute, Fujian Academy of Agricultural Sciences in Fuzhou (Fujian, China, 119°19′57″E, 26°7′47″N), and deposited in Hefei University of Technology (No. HFUTYSL003). Total genomic DNA including nuclear and organelle genome was extracted using CTAB protocol. A 150 bp paired-end library of total genomic DNA was constructed and sequenced, using HiSeq2000 platform (Illumina, USA) in Huitong Biotechnology (Shenzhen, China).
Totally, approximately 4.68 GB raw data was generated. The FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/) was employed to filter low-quality bases. Subsequently, based on sequence characteristics and similarity, the high-quality clean reads were used to assemble the cp genome using SPAdes v3.11.0 (Bankevich et al. Citation2012). The cp genome sequence was annotated using GeSeq (Tillich et al. Citation2017). The cp genome sequence of A. trifoliata with detailed annotations had been submitted to Genbank under accession number MN906448.
The cp genome is 157,952 bp in length with overall GC content 38.7%, which contains a large single-copy region (LSC) of 86,596 bp, a small single-copy region (SSC) of 19,058 bp, and two inverted repeat regions (IRA and IRB) of 26,149 bp. A total of 131 genes were annotated in the cp genome of A. trifoliata, including 86 protein-coding genes, 37 tRNA, and 8 rRNA genes.
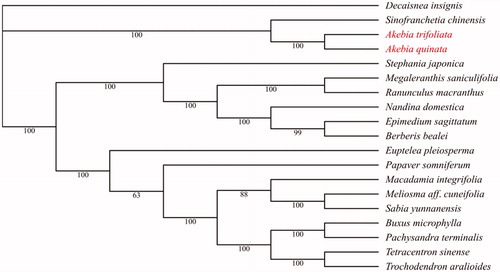
To better understand the phylogenetic relationship between A. trifoliata and other related plant species, the cp genome sequences of 19 plant species downloaded from GenBank were used to construct a phylogenetic maximum-likelihood tree using RaxML v8.2.9 (Stamatakis Citation2014), of which the bootstrap values were calculated using 1000 replicates. Phylogenetic analysis revealed A. trifoliata was clustered to Akebia quinata within Lardizabalaceae (), which was congruent with previous studies (Sun et al. Citation2016).
Figure 1. The phylogenetic tree based on the cp genome sequences of 19 plants. A. trifoliata and A. quinata are labeled in red. Bootstrap support values are given at the nodes. Accession number of 19 plants’ cp genome sequences is listed below: A. trifoliata (MN906448), A. quinata (NC_033913), Berberis bealei (NC_022457), Buxus microphylla (NC_009599), Decaisnea insignis (KY200671), Epimedium sagittatum (KU204899), Euptelea pleiosperma (KU204900), Macadamia integrifolia (NC_025288), Megaleranthis saniculifolia (NC_012615), Meliosma aff. cuneifolia (KU204901), Nandina domestica (NC_008336), Pachysandra terminalis (KU204904), Papaver somniferum (KU204905), Ranunculus macranthus (NC_008796), Sabia yunnanensis (KU204902), Sinofranchetia chinensis (NC_041488), Stephania japonica (KU204903), Tetracentron sinense (NC_021425) and Trochodendron aralioides (NC_021426).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that newly generated in this study is deposited in GenBank (www.ncbi.nlm.nih.gov), with accession number MN906448.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19 (5):455–477.
- Jiang Y, Du Y, Zhu X, Xiong H, Woo M, Hu J. 2012. Physicochemical and comparative properties of pectins extracted from Akebia trifoliata var. australis peel. Carbohydr Polym. 87 (2):1663–1669.
- Li L, Chen X, Yao X, Tian H, Huang H. 2010. Geographic distribution and resource status of three important Akebia species. Plant Sci J. 30(4):497–506.
- Li L, Xiaohong Y, Caihong Z, Xuzhong C, Hongwen H. 2010. Akebia: a potential new fruit crop in China. HortScience. 45 (1):4–10.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30 (9):1312–1313.
- Sun Y, Moore MJ, Zhang S, Soltis PS, Soltis DE, Zhao T, Meng A, Li X, Li J, Wang H. 2016. Phylogenomic and structural analyses of 18 complete plastomes across nearly all families of early-diverging eudicots, including an angiosperm-wide analysis of IR gene content evolution. Mol Phylogenet Evol. 96:93–101.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45 (W1):W6–W11.
- Wang J, Ren H, Xu QL, Zhou ZY, Wu P, Wei XY, Cao Y, Chen XX, Tan JW. 2015. Antibacterial oleanane-type triterpenoids from pericarps of Akebia trifoliata. Food Chem. 168:623–629.
- Xu QL, Wang J, Dong LM, Zhang Q, Luo B, Jia YX, Wang HF, Tan JW. 2016. Two new pentacyclic triterpene saponins from the leaves of Akebia trifoliata. Molecules. 21 (7):962.
- Zou S, Yao X, Zhong C, Zhao T, Huang H. 2019. Effectiveness of recurrent selection in Akebia trifoliata (Lardizabalaceae) breeding. Sci Hortic. 246:79–85.
