Abstract
Gila elegans, Hybognathus amarus, and Tiaroga cobitis (Family Cyprinidae, Order Cypriniformes) are endemic and endangered fishes in the southwestern United States. We present complete mitochondrial genomes for each species. Each mitochondrion consisted of 13 protein-coding genes, 2 ribosomal (rRNA) genes, 22 transfer RNA (tRNA) genes, and a single control region (D-loop), and gene order was consistent with other cyprinid fishes. Total genome lengths were 16,593 base pairs (bp) for G. elegans, 16,705 bp for H. amarus, and 16,802 for T. cobitis. The GC content in G. elegans and H. amarus was 44%, but higher in T. cobitis at 48%. Phylogenetic trees were generated to confirm relationships inferred via novel mitogenomes, and best-supported trees were consistent with previous research.
The vertebrate mitochondrial genome contains a highly conserved and well-understood gene composition and order, and yet can exhibit a rapid rate of sequence divergence between species and populations. These properties of the mitochondrial genome are useful for distinguishing and comparing recently diverged lineages (e.g. Ratnasingham and Hebert Citation2007). As such, mitochondrial genome studies have shed light on an array of ecological and evolutionary aspects of vertebrate life. Moreover, the mitochondrial genome is a valuable resource in conservation genetic research as it can be utilized for understanding changes in population dynamics and evolutionary history (Taguchi et al. Citation2015; Osborne et al. Citation2019) and in environmental DNA (eDNA) applications (e.g. Bronnenhuber and Wilson Citation2013; Dysthe et al. Citation2016). Fishes of the southwestern United States can benefit from comparative mtDNA research because many taxa have experienced precipitous declines and are subject to active management. Here, we assembled and annotated mitochondrial genomes for three western cyprinids: Bonytail (Gila elegans), Rio Grande Silvery Minnow (Hybognathus amarus), and Loach Minnow (Tiaroga cobitis). All three species are federally listed as endangered and have experienced major declines due to alterations to the natural hydrograph, habitat loss, and the introduction of non-native fishes (Sublette et al. Citation1990; Minckley and Marsh Citation2009). All three species have hatchery breeding programs, and have been stocked into natural habitats as part of recovery efforts.
Genomic DNA was isolated from caudal fin tissue from a single individual of each taxon. Gila elegans was collected from Southwestern Native Aquatic Resource and Recovery Center (Dexter, New Mexico, 33.194667, −104.350647) and H. amarus was collected from the Rio Grande, New Mexico (approximate locality: 34.213738, −106.885898). Tiaroga cobitis was collected from hatchery broodstock maintained at the Arizona Aquatic Research and Conservation Center that originated from the upper Gila River in New Mexico (approximate locality: 33.2284105, −108.255927). For all species, any remaining tissue and/or DNA isolates were deposited in the Museum of Southwestern Biology (MSB ACC2014-V.23, ACC1993-VIII:27 and ACC2018-X:30). Paired-end reads (150 bp) from a single sequencing lane (Illumina NextSeq 500; University of New Mexico) for each individual were quality trimmed via trimmomatic v. 0.36 (Bolger et al. Citation2014). Remaining paired reads were each baited and assembled using MITObim (Hahn et al. Citation2013) with the full mitogenomes of Gila robusta (Genbank Accession: NC008105.1), Hybognathus nuchalis (Genbank Accession: NC031567.1) and Rhinichthys cataractae (Genbank Accession: MG570448.1) used as bait sequences for G. elegans, H. amarus, and T. cobitis, respectively. Annotation of novel mitochondrial genomes was performed using the MitoFish pipeline (Iwasaki et al. Citation2013).
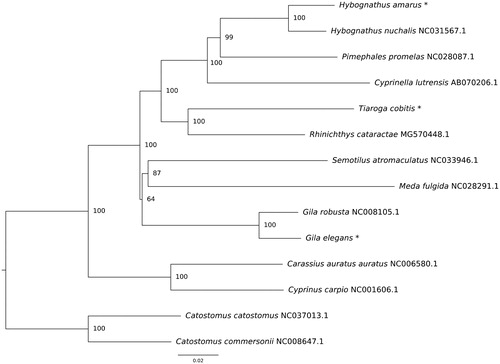
In all taxa, each mitochondrion consisted of 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and a D-loop control region. Gene order was identical to other cyprinid fishes. Complete genome lengths were 16,593 for G. elegans, 16,705 bp for H. amarus and 16,802 for T. cobitis. Minor genome length variation between species was the result of variation in the D-loop and origin of replication (Brown et al. Citation1993). Length of D-loop regions were 915 bp for G. elegans, 1042 bp for H. amarus, and 1135 bp in T. cobitis. Nucleotide content in G. elegans was 29% A, 27% T, 26% C, 18% G, H. amarus nucleotide content was 28% A, 29% T, 26% C, 17% G and nucleotide content for T. cobitis was 25% A, 27% T, 27% C, 21% G. Novel mitogenomes were imported into Mega7 (Kumar et al. Citation2016) and aligned with complete mitochondrial genomes of nine other cyprinid species and rooted with two representatives of the family Catostomidae. A neighbor-joining tree was constructed using composite maximum likelihood distances and node support was determined from 1000 bootstrap replicates. Results of the phylogenetic analysis revealed groupings consisted with previous research (; Schönhuth et al. Citation2018).
Acknowledgements
The authors gratefully acknowledge the expert assistance of George Rosenberg, Lijing Bu and Melissa Sanchez (UNM Core Facility) and Emily DeArmon (Museum of Southwestern Biology).
Disclosure statement
No potential conflict of interest was reported by the author(s). Research reported in this publication was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under award number P30 GM110907. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data availability statement
The mitochondrial genome sequences reported here are available on Genbank using the following accession numbers:
MT364325 (https://www.ncbi.nlm.nih.gov/nuccore/MT364325)
MT364326 (https://www.ncbi.nlm.nih.gov/nuccore/MT364326)
MT364327 (https://www.ncbi.nlm.nih.gov/nuccore/MT364327).
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Bronnenhuber, J.E. and Wilson, C.C., 2013. Combining species-specific COI primers with environmental DNA analysis for targeted detection of rare freshwater species. Conservation Genetics Res. 5(4):971–975.
- Brown JR, Beckenbach AT, Smith MJ. 1993. Intraspecific DNA sequence variation of the mitochondrial control region of white sturgeon (Acipenser transmontanus). Molecular Biology and Evolution. 10(2):326–41. https://doi.org/10.1093/oxfordjournals.molbev.a040007
- Dysthe JC, Carim KJ, Paroz YM, McKelvey KS, Young MK, Schwartz MK. 2016. Quantitative PCR assays for detecting loach minnow (Rhinichthys cobitis) and spikedace (Meda fulgida) in the southwestern United States. PLOS One. 11(9):e0162200.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Minckley WL, Marsh PC. 2009. Inland Fishes of the Greater Southwest: Chronicle of a Vanishing Biota. University of Arizona Press. Tucson, Arizona.
- Osborne MJ, Cordova SJ, Cameron AC, Turner TF. 2019. Isolation by elevation: mitochondrial divergence among sky island populations of Sacramento Mountain salamander (Aneides hardii). Conserv Genet. 20(3):545–556.
- Ratnasingham S, Hebert PD. 2007. BOLD: the barcode of life data system (http://www.barcodinglife.org). Mol Ecol Notes. 7(3):355–364.
- Schönhuth S, Vukić J, Šanda R, Yang L, Mayden RL. 2018. Phylogenetic relationships and classification of the Holarctic family Leuciscidae (Cypriniformes: Cyprinoidei). Mol Phylogenet Evol. 127:781–799.
- Sublette EJ, Hatch DM, Sublette M. 1990. The Fishes of New Mexico. University of New Mexico Press. Albuquerque, New Mexico.
- Taguchi M, King JR, Wetklo M, Withler RE, Yokawa K. 2015. Population genetic structure and demographic history of Pacific blue sharks (Prionace glauca) inferred from mitochondrial DNA analysis. Mar Freshwater Res. 66(3):267–275.