Abstract
Agave hybrid 11648 is the most widely cultivated agave variety for sisal fiber production around the world. In the present study, we have successfully sequenced the chloroplast genome of A. H11648. The complete chloroplast genome size is 157,274 bp in length with a GC content of 37.8%. The genome contains a large single copy region (LSC) of 85,896 bp, a small single copy region (SSC) of 18,230 bp, and a pair of inverted repeat regions (IR) of 26,574 bp. 121 genes are annotated in the chloroplast genome. The numbers of protein-coding, tRNA and rRNA genes are 99, 40 and 8, respectively. Phylogenetic tree reveals that A. H11648 is closely related to A. americana.
Sisal, an important kind of nature fiber with the properties of tough texture, high strength and friction resistance, has been widely used in navigation, automotive, papermaking, etc. (Li et al. Citation2000). As the main cultivar, A. H11648 ((A. amanuensis × A. angustifolia) × A. amaniensis) has been widely cultivated in tropical areas of Africa, South America and China (Robert et al. Citation2008). According to a previous study, a phylogenetic tree has been constructed with the partial chloroplast (cp) sequences of four agave species (Huang et al. Citation2018). However, the systematic position and phylogenetic relationship of A. H11648 still remain ambiguous at cp genome level. In the present study, we assembled the complete cp genome of A. H11648 with next-generation sequencing, with the aim to reveal the phylogenetic relationship of agave species.
The leaves of A. H11648 were collected the experimental field (22.90°N, 108.33°E) of Guangxi Subtropical Crops Research Institute, Nanning, China. The modified CTAB method was used to extract the total genomic DNA (Doyle and Doyle Citation1987). The specimen was deposited in Herbarium of Guangxi Subtropical Crops Research Institute (HGS-jm2018004). Total DNA was sent to Sangon Biotech (Shanghai, China) for library construction and next-generation sequencing. A total of 1.29 Gb short reads data was generated using Illumina HiSeq 2500 platform. The trimmed reads were selected for cp genome assembly by SPAdes software (Bankevich et al. Citation2012). The assembled cp genome was annotated by DOGMA (Wyman et al. Citation2004), which was corrected with Geneiousv11.0.3 (Kearse et al. Citation2012). The complete cp genome sequence was deposited in GenBank under the accession number MG642741.
The complete chloroplast genome size is 157,274 bp in length with a GC content of 37.8%. The genome contains a large single copy region (LSC) of 85,896 bp, a small single copy region (SSC) of 18,230 bp, and a pair of inverted repeat regions (IR) of 26,574 bp. 121 genes are annotated in the chloroplast genome. The numbers of protein-coding, tRNA and rRNA genes are 99, 40 and 8, respectively. Among these, ten genes (trnA-UGC, trnI-GAU, ndhB, ycf15, rpl2, trnK-UUU, atpF, rpoC1, trnL-UAA and trnV-UAC) contains one intron and three with two introns (ycf68, ycf3 and clpP).
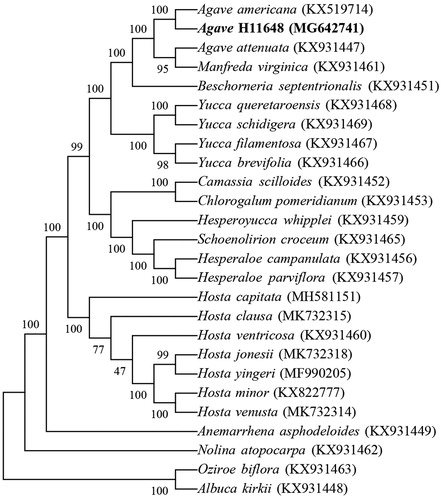
The cp genome sequences of 26 species were selected for phylogenetic analysis (23 species in Agavoideae and 3 species as outgroup including Albuca kirkii, Nolina atopocarpa and Oziroe biflora) (McKain et al. Citation2016; Lee et al. Citation2019). The result indicated that A. H11648 is closely related with A. americana (), which was consistent with our previous study (Huang et al. Citation2018). The cp genome sequences were aligned using MAFFT (Katoh and Standley Citation2013). The phylogenetic tree was constructed in MEGA7 software with the neighbor-joining method with 10000 bootstrap replicates (Kumar et al. Citation2016). This study will benefit future studies related to chloroplast in Agave genus.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
The data that support the findings of this study are fully available in GenBank (https://www.ncbi.nlm.nih.gov/nuccore/MG642741).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Huang X, Wang B, Xi J, Zhang Y, He C, Zheng J, Gao J, Chen H, Zhang S, Wu W, et al. 2018. Transcriptome comparison reveals distinct selection patterns in domesticated and wild Agave species, the important CAM plants. Int J Genom. 2018:1–12.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lee SR, Kim K, Lee BY, Lim CE. 2019. Complete chloroplast genomes of all six Hosta species occurring in Korea: molecular structures, comparative, and phylogenetic analyses. BMC Genomics. 20(1):833.
- Li Y, Mai YW, Ye L. 2000. Sisal fibre and its composites: a review of recent developments. Compos Sci Technol. 60(11):2037–2055.
- McKain MR, McNeal JR, Kellar PR, Eguiarte LE, Pires JC, Leebens-Mack J. 2016. Timing of rapid diversification and convergent origins of active pollination within Agavoideae (Asparagaceae). Am J Bot. 103(10):1717–1729.
- Robert ML, Lim KY, Hanson L, Sanchez-Teyer F, Bennett MD, Leitch AR, Leitch IJ. 2008. Wild and agronomically important Agave species (Asparagaceae) show proportional increases in chromosome number, genome size, and genetic markers with increasing ploidy. Bot J Linn Soc. 158(2):215–222.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20(17):3252–3255.