Abstract
The species of genus Hemipenthes can be easily identified by the following characters: wing with extensive infuscate pattern reaching hind margin basally and at least half as long as wing; antennal flagellum onion-shaped and not segmented; fore tibia smooth or with a few weak bristles. In this study, mitochondrial DNA (mtDNA) of the bee fly, Hemipenthes neimengguensis, has been annotated for the first time in the subfamily. The circular genome is 15,405 bp in length with an A + T content of 72.7%, and contains 13 protein-coding genes, 22 transfer RNAs, 2 ribosomal RNAs, and a partial control region (573 bp). All genes have similar locations and strands with that of other published Diptera. Phylogenetic analysis shows that Hemipenthes neimengguensis + Geron pallipilosus (Bombyliidae) were herein corroborated to be a sister group. And they have very closely related to Leptogaster longicauda and Satanas sp. which belong to the family Asilidae. Our results show the location of genus Hemipenthes in Bombyliidae and the location of the family in Brachycera, and provide data for further study of phylogeny in Diptera.
Genus Hemipenthes belongs to the tribe Villini of subfamily Anthracinae (Hull Citation1973) can be easily identified by the following characters: wing with extensive infuscate patterning reaching hind margin basally and at least half as long as wing; antennal flagellum onion-shaped and not segmented; fore tibia smooth or with a few weak bristles (Greathead and Evenhuis Citation1997; Yao and Yang Citation2008). The species H. neimengguensis Yao, Yang et Evenhuis is first report from China (Inner Mongolia) as a new species (Evenhuis and Greathead Citation2003; Yao et al. Citation2008; Yang et al. Citation2012). The biology of Hemipenthes species remains poorly known. Only the observation by author in Beijing in August 2007 found it’s like to congregated around flowers of the umbelliferous Petroselinum crispum and the adults of Hemipenthes like to rest in open places on the ground on the border between sunshine and shadow (Yao and Yang Citation2008). In this study, we assembled and characterized the mitochondrial genome of H. neimengguensis by high-throughput sequencing method for the first time. All the specimens or their DNA which studied in this research were deposited in the Entomological Museum of China Agricultural University, Beijing, China. The sequence is accessible from GenBank with the accession number MT043309.
The male adult sample of H. neimengguensis was collected from Lianhua Mountain, Kangle, Gansu, China in 2016 (N34°54′60″E 103°43′52″). The thorax muscle of the specimen was used to extract total genomic DNA. Vouchers consisting of the remaining bee fly were deposited in the Entomological Museum of China Agricultural University, Beijing, China (accession number: Bom-000113). The mitogenomes were amplified and sequenced as described in these studies. The sequence reads into contigs by using the program Geneious version 10.2.2 and BioEdit version 7.0.5.3 (Hall Citation1999). tRNA genes were identified by using the tRNAscan-SE (Lowe and Eddy Citation1997) and checked manually. Two rRNA genes and all protein-coding genes (PCGs) were identified by BLAST searches in NCBI (http://www.ncbi.nlm.nih.gov). Strand asymmetry was measured using the formulas: AT skew = [A − T]/[A + T] and GC skew = [G − C]/[G + C] (Perna and Kocher Citation1995).
The H. neimengguensis circular genome is more than 15,405 bp in length with an A + T content of 72.7%, and like other animals H. neimengguensis mitogenome is circular and contains 37 genes, including 13 PCGs, 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and a control region (could not sequence the entire control region in this study). The control region is more than 573 bp in length and located between srRNA and tRNAIle genes. All genes of H. neimengguensis mitogenomes have similar locations and strands with that of other published Brachycera. The composition of the mitogenome is biased toward A and T (72.7%), with A 38.8%, T 33.9%, C 16.7%, and G 10.5%. The A + T content of PCGs, tRNAs, and rRNAs are 71.1, 74.4, and 76.6%, respectively. The total length of all 13 PCGs in H. neimengguensis is 11,202 bp. Five PCGs (ND2, ATP8 ND5, and ND6) used ATT as start codon, six PCGs (COII, ATP6, COIII, ND4, ND4L, CytB) used ATG as start codon, one PCG used TCG as start codon, one PCG used TTG as start codon. Meanwhile, eight PCGs used the typical termination codons TAA, while two PCGs (ND1 and CytB) used TAG as termination codons, and three PCGs (COII, ND4, and ND5) stopped with the incomplete termination signal T.
The 22 tRNA genes size varies from 64 to 72 bp and all tRNAs could be folded into typical clover-leaf structures. The lengths of 12S and 16S rRNA genes are 793 and 1334 bp, respectively. In addition to the control region, the longest noncoding intergenic spacer is located between tRNAGlu and tRNAPhe with 19 nucleotides. Gene overlaps are also found at 11 gene junctions involving 41 nucleotides with the longest overlap (8 nucleotides) between tRNATrp and tRNACys, ND4, and ND4L.
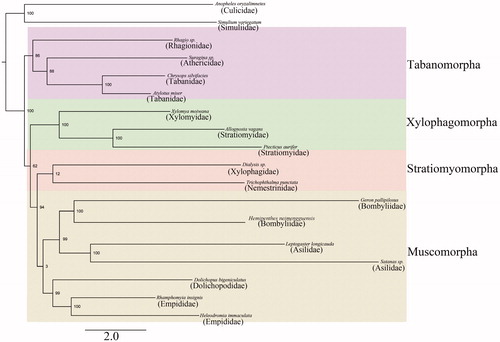
Based on published mitogenome sequences of Diptera species, phylogenetic analyses were performed with respect to the nucleotide sequences of 13 PCGs and 2 rRNAs. Phylogenetic trees () based on the mitogenome data were inferred by both Maximum likelihood (ML) and Bayesian methods (BI). The species of mitochondrial genome sequences from GenBank accession no. were as following: Anopheles oryzalimnetes-NC_030715; Simulium variegatum-NC_033348; Rhagio sp.-KT225298; Suragina sp.-KT225301; Chrysops silvifacies-KT225292; Atylotus miser-KT225291; Xylomya moiwana-KT225302; Ptecticus aurifer-KT225297; Dialysis sp.-KT225293; Trichophthalma punctate-NC_008755; Geron pallipilosus-MG732929; H. neimengguensis-MT043309; Leptogaster longicauda-KT225296; Satanas sp.-KT225300; Dolichopus bigeniculatus-KT225294; Rhamphomyia insignis-KT225299; Heleodromia immaculate-KT225295. The results phylogenetic analysis shows that all infraorders of Brachycera are monophyletic, and it shows H. neimengguensis + Geron pallipilosus (Bombyliidae) was herein corroborated to be a sister group. And they have very closely related to Leptogaster longicauda and Satanas sp. which belong to the family Asilidae. In this monophyletic bombyliidae and Asilidae was assigned to be a sister group, and Dolichopodidae and Empididae was herein corroborated to be a sister group, this result is supported by previous studies based on morphological characters (Yeates et al. Citation2007; Lambkin et al. Citation2013).
In summary, the mitochondrial genome of H. neimengguensis (Bombyliidae) will provide important molecular data for further evolutionary analysis for the phylogeny status of the family, the genome data will also provide fundamental information for the conservation, pollinating, and natural enemy of this important species.
Acknowledgments
We are very grateful to Mr. Xuankun Li (Canberra) and Mr. Liang Wang for their help in phylogenetic analysis based on mitogenome sequences.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available in Gang Yao at [https://pan.baidu.com/s/1dURoaP5c—pprPnsYXR1g. with the code: r3tp].
Additional information
Funding
References
- Evenhuis NL, Greathead DJ. 2003. World catalog of bee flies (Diptera: Bombyliidae). Zootaxa. 300(1):1.
- Greathead DJ, Evenhuis NL. 1997. Family Bombyliidae. In: Papp L, Darvas B, editors. Manual of palaearctic diptera. Vol. 2. Budapest: Science Herald Press; p. 487–512.
- Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. Vol. 41; p. 95–98. https://pan.baidu.com/s/1dURoaP5c—pprPnsYXR1g. with the code: r3tp
- Hull FM. 1973. Bee flies of the world. The genera of the family Bombyliidae. Bull United States Nat Mus. 286:1–687.
- Lambkin CL, Sinclair BJ, Pape T, Courtney GW, Skevington JH, Meier R, Yeates DK, Blagoderov V, Wiegmann BM. 2013. The phylogenetic relationships among infraorders and superfamilies of Diptera based on morphological evidence. Syst Entomol. 38(1):164–179.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at four-fold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41(3):353–358.
- Yang D, Yao G, Cui WN. 2012. Bombyliidae of China. Beijing, China: China Agricultural University.
- Yao G, Yang D. 2008. Two new species of Hemipenthes Loew, 1869 from Oriental China (Diptera: Bombyliidae. Zootaxa. 1689(1):63–68.
- Yao G, Yang D, Evenhuis NL. 2008. Species of Hemipenthes Loew, 1869 from Palaearctic China (Diptera: Bombyliidae. Zootaxa. 1870(1):1–23.
- Yeates DK, Wiegmann BM, Courtney GW, Meier R, Lambkin C, Pape T. 2007. Phylogeny and systematic of Diptera: two decades of progress and prospects. Zootaxa. 1668(1):565–590.