Abstract
For the first time, we illuminate the complete mitochondrial genome (mitogenome) sequence of the Paradiplospinus antarcticus, which is 16,988 bp in size and contains 13 protein-coding (PCGs), 2 rRNA genes, 22 tRNA genes, and one control region.The base composition of the mitogenome is 26.08% A, 26.77% T, 28.46% C and 18.69% G. Here, we selected 11 genera of species from the mostly monotypic snake mackerel family, including representative Antarctic Paradiplospinus antarcticus that have been identified, and constructed phylogenetic trees to better study the snake mackerel family.
Fishes of the family Gempylidae (snake mackerels) are widely distributed in deeper tropical and subtropical waters (Nakamura and Parin Citation1993). There are 24 species in 16 genera in the world (Nelson et al. Citation2016). However, the question of the classification status of the snake mackerel family in the Scombroidei has been puzzling for some time (Johnson Citation1986).One of the snake mackerels, Paradiplospinus antarcticus, was discovered and named by Andriashev in 1960 (Andriashev Citation1960). Paradiplospinus antarcticus and Paradiplospinus gracilis are similar in morphology, with some overlap in distribution. Some species collected from the Southern Ocean are directly referred to as ‘Paradiplospinus gracilis’, and it is controversial whether the two are the same.
The valuable samples (Deposit Number: Fish-1-Antarctic 36) were collected in the Southern Ocean (63°01′S, 058°05′W) and stored in the Fish Collection in the Institute of Zoology, Chinese Academy of Sciences, Beijing, China (IOZCAS). Total genomic DNA was isolated using Easy Pure@Genomic DNA Kit according to the manufacturer’s instructions (TransGenBiotech Co, Beijing, China). The mitochondrial genome sequence of Paradiplospinus antarcticus was determined and annotated by Biomarker Technologies Co, LTD, Beijing, China. Phylogenetic trees were reconstructed based on the 13protein-coding genes (PCGs) of 12 snake mackerels and out-group taxa using IQ-TREE web server with the TPM2 + F + I + G4 model (Trifinopoulos et al. Citation2016).
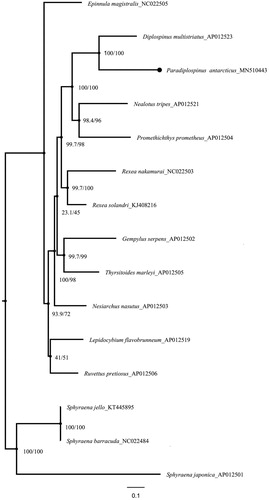
The complete and circular mitochondrial genome is 16,988 bp in size, it has been deposited in NCBI and Genbank (accession: MN510443) with an AT bias of 52.85%, and includes 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), 2 ribosomal RNA genes (12S rRNA and 16S rRNA), and one control regions (CR). There are ND6 subunit gene and eight tRNAs on the L-strand, and the other 28 genes on the H-strand. Thirteen PCGs are 12,869 bp and start with typical ATN codons except COX1 with GTG; The TAG was found as the stop codon in COX1, ND3, ND5 and ND6, the TAA was found as the stop codon in ND1, ND2, ATP8, ATP6, COX3 and ND4L, while the incomplete codon T was found as the stop codon in COX2, ND4 and CYTB; The 22 tRNA genes range in size from 66 bp in tRNACys to 75 bp in tRNALeu and tRNA Lys, the12S and 16S rRNA are 956 and 1687 bp, respectively. The result showed that most of the nodes in the tree graph have higher SH-aLRT and ultrafast bootstrap values (). Besides, Paradiplospinusantarcticus and Diplospinus multistriatus, are closely clustered to support previous results based on morphological studies (Jondeung and Karinthanyakit Citation2010).
Data availablity statement
The data that support the findings of this study are openly available in [NCBI], reference number [https://www.ncbi.nlm.nih.gov/nuccore/MN510443.1/].
Acknowledgments
We thank Captain Yanping Zhao, Chief Engineer Rong Huang, Chief Officer Xude Zhang and all crew aboard the XUE LONG 2 icebreaker for security and logistical support during the 36th Chinese National Antarctic Research Expedition.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Andriashev AP. 1960. Families of fishes new to the Antarctic. 1. Paradiplospinus antarcticus gen. et sp. n. (Pisces, Trichiuridae). Zoologicheskii Zhurnal. 39(2):244–249.
- Johnson GD. 1986. Scombroid phylogeny: an alternative hypothesis. Bull Mar Sci. 39:1–41.
- Jondeung A, Karinthanyakit W. 2010. The complete mitochondrial DNA sequence of the short mackerel (Rastrelliger brachysoma), and its phylogenetic position within Scombroidei, Perciformes. Mitochondrial DNA. 21(2):36–47.
- Nakamura I, Parin NV. 1993. FAO species catalogue, Snake mackerels and cutlassfish of the world (Families Gempylidae and Trichiuridae). FAO Fish Synos. 125:20–60.
- Nelson JS, Grande TC, Wilson M. 2016. Fishes of the world. 5th ed. New York (NY): Wiley and Sons; p. 415.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.