Abstract
The complete mitochondrial genome of Notothenia rossii was obtained using PacBio Sequel long-read sequencing platform. The mitogenome of N. rossii was circular form and 18,274 bp long, which consists of 13 protein-coding genes, 24 tRNAs, 2 rRNAs, and non-coding control region. Particularly, we found duplicated tRNAThr and tRNAPro in addition to the typical 22 tRNAs. The phylogenetic tree revealed that N. rossii was most closely related to N. coriiceps among species in the Nototheniidae clade within the suborder Notothenioidei.
Notothenia rossii Richardson, 1844 is a species belonging to the family Nototheniidae, also called the common name marbled rockcod. Like other notothenid fish, N. rossii widely distributes in the Southern Ocean and around Antarctica, although the population has been decreased by overfishing due to commercially importance in the fisheries (Barrera-Oro and Marschoff Citation2007). There are 7 recognized species in the genus Notothenia according to the FishBase (Froese and Pauly Citation2019). Up to now, mitochondrial and whole genome sequences in the genus Notothenia have been reported for only one species of N. coriiceps (Shin et al. Citation2014; Oh et al. Citation2016,). Here, we provide the complete mitochondrial genome of N. rossii (GenBank accession No. MT192936) and the results of the phylogenetic analysis with Antarctic fish species.
The sample of N. rossii was collected from the sea near Barton Peninsula, King George Island, West Antarctica (62°14′S, 58°47′W). The specimen was deposited at the Earth Biocollection in the Division of Biotechnology, Korea University with accession number KAN0007030. The genomic DNA was extracted by phenol/chloroform method and then g-TUBE (Covaris, CA, USA) shearing device and BluePippin system (Sage Science, MA, USA) were used for preparing 20 kb size-selected templates. The SMRTbell library preparation and sequencing were performed using PacBio Sequel platform according to the manufacturer’s protocol (Pacific Biosciences, CA, USA). The quality and quantity for library were checked with Fragment analyzer (Agilent Technologies, CA, USA) and Qubit 2.0 Fluorometer (Invitrogen, Life Technologies, CA, USA), respectively. PacBio subreads for mitochondrial genome were filtered out using partial sequences of 16 s rRNA and COX1 genes. De novo assembly was conducted by CANU assembler (Koren et al. Citation2017) and the assembled genome was annotated via MITOS web server (Bernt et al. Citation2013).
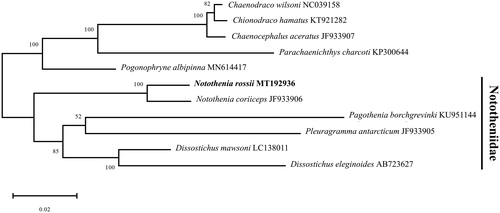
The complete mitochondrial genome of N. rossii (GenBank number: MT192936) was 18,274 bp in length, including 13 protein-coding genes, 24 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs) and non-coding control region. In particular, duplicated tRNAThr and tRNAPro are present in addition to the 22 tRNAs commonly found in fishes. The base composition of A + T (54.10%) was higher than the G + C (45.90%) in the mitogenome. Most of the protein coding genes have typical start codon (ATG) except for COX1, ATP8, and ATP6 genes (GTG). Three types of stop codons (TAA, TAG, and T(AA)) were identified. The phylogenetic relationships of N. rossii were analyzed with 10 Antarctic species in the suborder Notothenioidei using 13 protein-coding genes (). Maximum Likelihood (ML) tree was built with 500 bootstrap replications and JTT matrix-based model (Jones et al. Citation1992) by MEGA X software (Kumar et al. Citation2018). The tree showed that N. rossii was first clustered with N. coriiceps, and they were grouped with species belonging to Nototheniidae (Dissostichus eleginoides, D. mawsoni, Pagothenia borchgrevinki and Pleuragramma antarcticum). This result provides fundamental data to further our understanding of the evolutional relationship of Antarctic fishes in the Southern ocean.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI under the accession MT192936 (https://www.ncbi.nlm.nih.gov/nuccore/MT192936.1/).
Additional information
Funding
References
- Barrera-Oro ER, Marschoff ER. 2007. Information on the status of fjord Notothenia rossii, Gobionotothen gibberifrons and Notothenia coriiceps in the lower South Shetland Islands, derived from the 2000–2006 monitoring program at Potter Cove. CCAMLR Science. 14:83–87.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Froese, R. & Pauly, D., Editors. 2019. FishBase. World Wide Web electronic publication. www.fishbase.org. version (12/2019).
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Bioinformatics. 8(3):275–282.
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Oh JS, Ahn DH, Lee J, Choi J, Chi YM, Park H. 2016. Complete mitochondrial genome of the Antarctic bullhead notothen, Notothenia coriiceps (Perciformes, Nototheniidae). Mitochondrial DNA Part A. 27(2):1407–1408.
- Shin SC, Ahn DH, Kim SJ, Pyo CW, Lee H, Kim M-K, Lee J, Lee JE, Detrich HW, Postlethwait JH, et al. 2014. The genome sequence of the Antarctic bullhead notothen reveals evolutionary adaptations to a cold environment. Genome Biol. 15(9):468.