Abstract
We reported the complete chloroplast genome sequences of Rhus potaninii which was characterized by de novo assembly with Illumina sequencing data. The size of R. potaninii complete chloroplast genome is 159,620 bp in length and includes a large single copy region of 87,722 bp, a small single copy region of 18,948 bp, and a pair of inverted repeats of 26,475 bp. Its GC content is 37.9%. A total of 133 genes were predicted, including 86 protein-coding genes, 8 rRNA genes, 37 tRNA genes, and 2 pseudogenes. Maximum-likelihood (ML) phylogenetic tree indicates that R. potaninii is sister to R. chinensis.
Keywords:
The deciduous tree Rhus potaninii is 5–8 m tall. It grows on the hill or mountain forests at an altitude of 900–2500 m. The R. potaninii belongs to Anacardiaceae family of Sapindales order. The Galla chinensis, induced by infection with Gallnut Aphid on R. chinensis, R. potaninii, and R. punjabensis, is a traditional Chinese medicine (National Pharmacopoeia Committee Citation2010). It is known to have analgesic and anti-inflammatory effects (Sun et al. Citation2018). In recent years, Galla chinensis has attracted the attention of drug researchers at home and abroad because of its unique advantages such as strong pharmacological effect, extensive sources, and strong selectivity (Karimi-Khouzani et al. Citation2017). Although there are many pharmacology related research of it, there are few researches on its genome. Therefore, our research provides a basis for the study of the coevolution of Gallnut Aphid and host tree and the further study on phylogenetic evolution of R. potaninii.
Fresh leaves of R. potaninii were collected from Danfeng County, Shaanxi Province (33°36′33.52″N, 110°14′04.89″E). The specimen was saved in Herbarium of Institute of Medicinal Plant Development (voucher: Pan0101). Its total genomic DNA was extracted using QIAGEN DNeasy Plant Mini Kit and the whole genome was sequenced on Illumina Hiseq 2000 platform. Using SOAPdenovo 2 (Luo et al. Citation2012) and SSPACE (Boetzer et al. Citation2011) to assemble clean data into a complete chloroplast genome.
The total length of chloroplast genome of R. potaninii was 159,620 bp (Genbank accession number: MT230556) with 37.9% GC content. The genome consists of four distinct parts, including a large single copy region of 87,722 bp, a small single copy region of 18,948 bp, and a pair of inverted repeats of 26,475 bp. In the genome, 86 protein-coding genes, 8 rRNA genes, and 2 pseudogenes were identified by CPGAVAS (Liu et al. Citation2012), DOGMA (http://dogma.ccbb.utexas.edu/) (Wyman et al. Citation2004) and BLAST search (Cui et al. Citation2019), and 37 tRNA genes were annotated by tRNAscan-SE (Schattner et al. Citation2005).
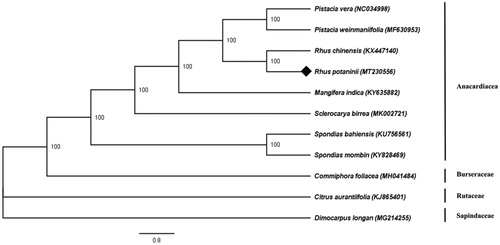
Phylogenetic analysis of the complete chloroplast genome sequence of R. potaninii and 10 species was performed through a maximum likelihood (ML) analyses with 1000 bootstrap replicates (), 10 species of which belonging to Sapindales order. The ML tree showed that R. potaninii was sister to R. chinensis with 100% support value, belonging to the Anacardiaceae family.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MT230556, accession number MT230556.
Additional information
Funding
References
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 27(4):578–579.
- Cui Y, Chen X, Nie L, Sun W, Hu H, Lin Y, Li H, Zheng X, Song J, Yao H. 2019. Comparison and phylogenetic analysis of chloroplast genomes of three medicinal and edible Am-omum species. IJMS. 20(16):4040–4054.
- Karimi-Khouzani O, Heidarian E, Amini SA. 2017. Anti-inflammatory and ameliorative effects of gallic acid on fluoxetine-induced oxidative stress and liver damage in rats. Pharmacol Rep. 69(4):830–835.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1(1):18.
- National Pharmacopoeia Committee. 2010. Pharmacopoeia of People’s Republic of China. Beijing: Chemical Industry Press.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689.
- Sun K, Song X, Jia R, Yin Z, Zou Y, Li L, Yin L, He C, Liang X, Yue G, et al. 2018. Evaluation of analgesic and anti-inflammatory activities of water extract of Galla chinensis in vivo models. Evid Based Complement Alternat Med. 2018:1–7.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.