Abstract
One complete mitochondrial genome (mitogenomes) was determined for Melanochlamys sp. (Cephalaspidae: Aglajidae). The mitochondrial genome size was 13,795 bp. The sequence contains two ribosomal RNA genes (rrnL and rrnS), 20 tRNA genes, and 12 protein-coding genes (PCGs). The A + T content of the complete mitochondrial genome sequence was 68.2%. The base composition showed a tendency of high AT. The resulted maximum-likelihood (ML) tree of Opisthobranchia supported that genetic differences between Melanochlamys sp. with other species of Cephalaspidae, so that there is a well-defined separation of clades.
Keywords:
Melanochlamys Cheeseman, 1881, is a relatively small group of aglajid sea slugs (Gastropoda, Cephalaspidea, Aglajidae), typically found crawling on soft bottoms. Rudman (Citation1972, Citation1974) established the diagnostic characteristics for Melanochlamys sp. and described the anatomy of several species in great detail. Recent molecular phylogenies of aglajid sea slugs (Camacho-García et al. Citation2014) indicate that Melanochlamys sp. is monophyletic. Here, we sequenced the mitochondrial genomes (mitogenomes) of a Chinese new species of the genus Melanochlamys. This species was collected in the culture pond of Apostichopus japonicus and considered as a new enemy of A. japonicus. In order to provide a theoretical basis and scientific guidance for the Melanochlamys sp., mitochondrial gene composition, biological evolution, and molecular identification were studied.
Melanochlamys sp. was collected from Dongying Shandong Province of China (37.60 N, 118.96E) in 2019. The specimen was deposited into Fishery Barcode Database of China (YSF-ZW-4635). Genomic DNA was extracted using OMEGA Mollusk DNA Kit. Paired-end sequencing (2 × 150 bp) was performed in an Illumina NextSeq sequencer. The reads were assembled using ABySS v2.0.2 (http://www.bcgsc.ca/platform/bioinfo/software/abyss). Subsequently, we annotated the assembled mitochondrial genome via DOGMA (Wyman et al. Citation2004).
The complete mitogenome sequences was 13,795 bp in length submitted in GenBank (Accession No. MT370509). The complete mitogenome contained two ribosomal RNA genes (rrnL and rrnS), 20 tRNA genes, and 12 protein-coding genes (PCGs). The length, the position, the start and stop codons of each PCG, the anticodon of each tRNA gene, and the coding strand are summarized in . The A + T content of the complete mitochondrial genome sequence was 68.2%. The base composition showed a tendency of high AT.
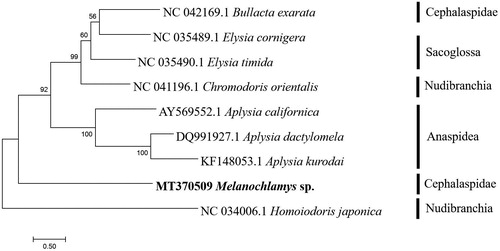
The phylogenetic tree was constructed by maximum-likelihood (ML) method using the MEGA 7 software. The result supported that all the species of Anaspidea formed a monophyletic clade with 100% support (). Melanochlamys sp. has a distant relationship with Bullacta exarate of Cephalaspidae. This suggests that genetic differences between Melanochlamys sp. with other species of Cephalaspidae, so that there is a well-defined separation of clades. Camacho-García et al. (Citation2014), in a comprehensive phylogenetic analysis of the Aglajidae, were able to resolve Melanochlamys sp. as a monophyletic group.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank(https://www.ncbi.nlm.nih.gov/) with reference number [MT370509].
Additional information
Funding
References
- Camacho-García YE, Ornelas-Gatdula E, Gosliner TM, Valdés Á. 2014. Phylogeny of the family Aglajidae (Pilsbry, 1895) (Heterobranchia: Cephalaspidea) inferred from mtDNA and nDNA. Mol Phylogenet Evol. 71:113–126.
- Rudman WB. 1972. On Melanochlamys Cheeseman, 1881, a genus of the Aglajidae (Opisthobranchia: Gastropoda). Pacific Sci. 26:50–62.
- Rudman WB. 1974. A comparison of Chelidonura, Navanax and Aglaja with other genera of the Aglajidae (Opisthobranchia: Gastropoda). Zool J Linn Soc. 54:185–212.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.