Abstract
Cymbidium serratum is a dominant species in the large orchid family with beautiful flowers, thick petals, and long flowering periods, and has a long history of cultivation in Southwest China. However, its wild resources have been threatened with extinction due to environmental degradation and artificial exploitation. In this study, the complete chloroplast genome of C. serratum was obtained through Illumina sequencing. The size of chloroplast genome of C. serratum is 149,998 bp, including large single-copy (LSC) and small single-copy (SSC) regions over 84,854 bp and 13,926 bp, respectively, and two inverted repeats (IRs) of 25,609 bp. The total GC content was 37.11%. The chloroplast genome contains 129 genes, including 83 protein-coding genes, 8 rRNA genes, and 38 tRNA genes. The maximum-likelihood phylogenetic tree indicated that C. serratum is a sister species with the clade composed of C. faberi and C. goeringii. The complete chloroplast genome of C. serratum will contribute to protecting this highly endangered species, and provide genetic information about genetic diversity and sustainable use.
Cymbidium serratum (Orchidaceae) is one of the eight traditional Chinese orchids. It is mainly distributed in the Yunnan Plateau, also in the mountainous areas with high altitudes at the junction of Sichuan, Guizhou, and Yunnan (Zhaoyang et al. Citation2010; Dagui Citation2011). Cymbidium serratum has extremely colorful flowers, which can be attributed to factors such as complex terrain, diverse climate types, strong ultraviolet rays in high altitude areas (Yufang Citation2005; Weiqing Citation2014). Cymbidium serratum is very limited in resources because of its narrow growing area (Yufang Citation2005). In recent years, due to the deterioration of the ecological environment and repeated exploitation, the wild resources of C. serratum have been Endangered (Huixian et al. Citation2010). In view of its threatened situation, it is urgent to strengthen the resource conservation and genetic breeding research of the species. Since the chloroplast genome sequence is inherited from matrilineal plants in most angiosperms and contains a great quantity of valuable genetic information (Daniell et al. Citation2016; Gitzendanner et al. Citation2018), it is widely used in species conservation, genome evolution and phylogenetic studies at various species levels (Barrett et al. Citation2013; Turmel et al. Citation2019). In this study, the complete chloroplast genome of C. serratum is reported providing valuable genetic information for further research on genetic diversity and conservation of this highly Endangered species.
The fresh leaves of C. serratum were collected from Bijie (105°37′59.16″E; 27°29′14.98″N), Guizhou Province, China. Voucher specimen was deposited at Orchid Resource Nursery of Zhejiang Agriculture and Forestry University (specimen code ZAFU20140310). The total genomic DNA was extracted by modified CTAB method (Fu et al. Citation2017) and sequenced by NovaSeq platform (Illumina, USA). The complete cp genome was assembled using SPAdes v3.10.1 (Bankevich et al. Citation2012) and genome annotation was performed with Geneious ver. 10.1 (Kearse et al. Citation2012). The whole chloroplast genome sequence of C. serratum has been submitted to GenBank (accession number MT273089).
The size of chloroplast genome of C. serratum is 149998 bp, including large single-copy (LSC) and small single-copy (SSC) regions over 84,854 and 13,926 bp, respectively, and two inverted repeats (IRs) of 25,609 bp. The total GC content is 37.11%, and the GC content of LSC, SSC, and IR are 34.46, 29.63, and 43.53%, respectively. The chloroplast genome of C. serratum contains 129 genes, including 83 protein-coding genes, 8 rRNA genes, and 38 tRNA genes. There are 19 genes duplicated in the IRs, and the remaining 110 genes are unique genes. Thirteen genes contain one intron, while three genes have two introns.
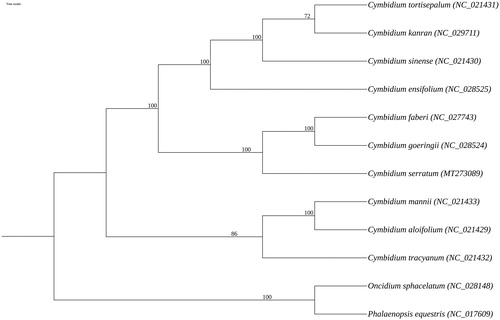
To confirm the phylogenetic status of C. serratum, phylogenetic analysis was performed based on the complete chloroplast genomes of nine Cymbidium species and two Orchidaceae species as outgroups downloaded from GenBank. MAFFT v7.388 (Nakamura et al. Citation2018) was applied for multisequence alignment, and the aligned sequences were employed to construct the maximum-likelihood phylogenetic tree using RAxML v8.2.10 (Stamatakis Citation2014) with 1000 bootstrap. The ML phylogenetic analysis manifests that C. serratum is a sister species with the clade composed of C. faberi and C. goeringii ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov, GenBank Accession Number: MT273089.
Additional information
Funding
References
- Barrett CF, Davis JI, Leebens-Mack J, Conran JG, Stevenson DW. 2013. Plastid genomes and deep relationships among the commelinid monocot angiosperms. Cladistics. 29(1):65–87.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Dagui L. 2011. Cymbidium serratum ‘Hongheyinxiang’: beautiful in color and appearance. Flower Plant Penjing. (10):32.
- Daniell H, Lin C-S, Yu M, Chang W-J. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134–162.
- Fu Z, Song J, Jameson PE. 2017. A rapid and cost effective protocol for plant genomic DNA isolation using regenerated silica columns in combination with CTAB extraction. J Integr Agric. 16(8):1682–1688.
- Gitzendanner MA, Soltis PS, Yi T-S, Li D-Z, Soltis DE. 2018. Plastome phylogenetics: 30 years of inferences into plant evolution. In: Chaw S-M, Jansen RK, editors. Advances in botanical research. New York: Academic Press; p. 293–313.
- Huixian P, Hongming Y, Jun M, Yingjie L, Yingbai S. 2010. Seed germination characteristics of Cymbidium goeringii, C. goeringii var. goeringii, C. tortispalum and C. goeringii var. longibracteatum. J West China For Sci. 39(3):56–60.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 34(14):2490–2492.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Turmel M, Lopes dos Santos A, Otis C, Sergerie R, Lemieux C. 2019. Tracing the evolution of the plastome and mitogenome in the chloropicophyceae uncovered convergent tRNA gene losses and a variant plastid genetic code. Genome Biol Evol. 11(4):1275–1292.
- Weiqing Y. 2014. The famous orchid: Cymbidium serratum ‘Honghehong. Flowers. (10):32.
- Yufang Y. 2005. Cultivation and management technology of Cymbidium serratum. Appl Technol Rural Areas. (3):27.
- Zhaoyang B, He J, Bi Z, Dong W, Yan H. 2010. Asepsis sowing and in vitro propagation of Cymbidium serratum (Schltr) Y. S. Wu et S. C. Chen. Plant Physiol Commun. 46(12):1259–1260.