Abstract
The complete chloroplast genome sequence of Populus adenopoda was characterized from Illumina pair-end sequencing. The chloroplast genome of P. adenopoda was 155,533 bp in length, containing a large single-copy region (LSC) of 84,818 bp, a small single-copy region (SSC) of 16,845 bp, and two inverted repeat (IR) regions of 26,935 bp. The overall GC content is 30.7%, while the corresponding values of the LSC, SSC, and IR regions are 34.5%, 30.5%, and 42.3%, respectively. The genome contains 131 complete genes, including 86 protein-coding genes (62 protein-coding gene species), 37 tRNA genes (29 tRNA species) and 8 rRNA genes (4 rRNA species). The neighbour-joining phylogenetic analysis showed that P. adenopoda and Populus alba clustered together as sisters to other Populus species.
Introduction
The genus Populus in the family Salicaceae includes over 100 tree species distributed in the world, with 53 species in China. Populus species, collectively known as poplar, are known for their commercially and ecologically forest trees. Populus adenopoda, an endemic species, occurs only at high altitudes in Sichuan of SW China but is often confused with alba, and the systematics of the species is unclear (Hou et al. Citation2018). Populus adenopoda has high ecological and economic value with high levels of intraspecific genetic diversity. Populus adenopoda has wide geographic distribution, high intraspecific polymorphism, adaptability to different environments, combined with a relatively small genome size. Consequently, P. adenopoda represents an excellent model for understanding how different evolutionary forces have sculpted the variation patterns in the genome during the process of population differentiation and ecological speciation (Neale and Antoine Citation2011). Moreover, we can develop conservation strategies easily when we understand the genetic information of P. adenopoda. In the present research, we constructed the whole chloroplast genome of P. adenopoda and understood many genome variation information about the species, which will be beneficial for population genetics studies of P. adenopoda.
The fresh leaves of P. adenopoda were collected from Zhejiang (30°12′N, 119°29′EThey were silica-dried and taken to the laboratory for DNA extraction. The voucher specimen (XXY001) was laid in the Herbarium of China West Normal University and the extracted DNA was stored in the –80 °C refrigerator of the Key Laboratory of Southwest China Wildlife Resources Conservation. We extracted total genomic DNA from 25 mg silica-gel-dried leaf using a the modified CTAB method (Doyle Citation1987). The whole-genome sequencing was then conducted by Biodata Biotechnologies Inc. (Hefei, China) with Illumina Hiseq platform. The Illumina HiSeq 2000 platform (Illumina, San Diego, CA) was used to perform the genome sequence. We used the software MITObim 1.8 (Hahn et al. Citation2013) and metaSPAdes (Nurk et al. Citation2017) to assemble chloroplast genomes. We used P. tremuloides (GenBank: MN561844) as a reference genome. We annotated the chloroplast genome with the software DOGMA (Wyman et al. Citation2004), and then corrected the results using Geneious 8.0.2 (Campos et al. Citation2016) and Sequin 15.50 (http://www.ncbi.nlm.nih.gov/Sequin/).
The complete chloroplast genome of P. adenopoda (GenBank accession number MT482539) was characterized from Illumina pair-end sequencing. The chloroplast genome of P. adenopoda was 155,533 bp in length, containing a large single-copy region (LSC) of 84,818 bp, a small single-copy region (SSC) of 16,845 bp, and two inverted repeat (IR) regions of 26,935 bp. The overall GC content is 30.7%, while the corresponding values of the LSC, SSC, and IR regions are 34.5%, 30.5%, and 42.3%, respectively. The genome contains 131 complete genes, including 86 protein-coding genes (62 protein-coding gene species), 37 tRNA genes (29 tRNA species), and 8 rRNA genes (4 rRNA species).
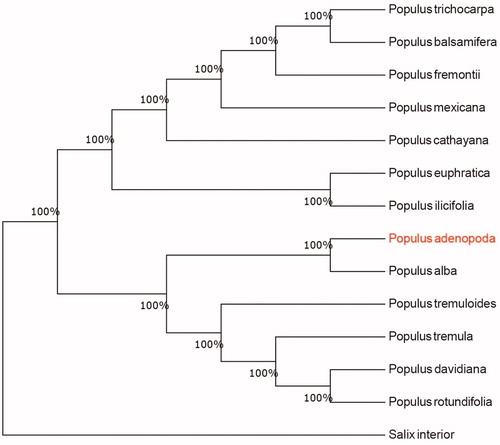
We used the complete chloroplast genomes sequence of P. adenopoda and 12 other related species of Populus and Salix interior as outgroup to construct the phylogenetic tree. The 14 chloroplast genome sequences were aligned with MAFFT (Katoh and Standley Citation2013), and then the neighbour-joining tree was constructed using MEGA 7.0 (Kumar et al. Citation2016). The results confirmed that P. adenopoda was clustered with P. alba ().
Figure 1. Neighbour-joining (NJ) analysis of P. adenopoda and other related species based on the complete chloroplast genome sequence. Genbank accession numbers: P. davidiana (KP861984), P. koreana (MN864049), P. yunnanensis (KP729176), P. euphratica (KJ624919), P. adenopoda (MT482539), P. rotundifolia (KX425853), P. cathayana (KP929175), P. balsamifera (KJ664927), P. ilicifolia (NC031371), P. trichocarpa (EF489041), P. fremontii (KJ664926), P. tremuloides (MN561844) and Salix interior (NC024681).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MT482539.
Additional information
Funding
References
- Campos FS, Kluge M, Franco AC, Giongo A, Valdez FP, Saddi TM, Brito WMED, Roehe PM. 2016. Complete genome sequence of porcine parvovirus 2 recovered from swine sera. Genome Announc. 4(1):e0162701615.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Hou Z, Wang Z, Ye Z, Du S, Liu S, Zhang J. 2018. Phylogeographic analyses of a widely distributed Populus davidiana: Further evidence for the existence of glacial refugia of cool-temperate deciduous trees in northern East Asia . Ecol Evol. 8(24):13014–13026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Neale DB, Antoine K. 2011. Forest tree genomics: growing resources and applications. Nat Rev Genet. 12(2):111–122.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
