Abstract
We determined the mitogenome sequence of Jesogammarus (Jesogammarus) hinumensis Morino, Citation1993, which is the first complete mitogenome sequence in the family Anisogammaridae Bousfield, 1977. The complete mitogenome of J. (J.) hinumensis was 15,186 bp in length with the typical 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and a control region (CR). The gene order of J. (J.) hinumensis was in accordance with the typical pan-crustacean ground pattern. A maximum-likelihood tree constructed using 25 eumalacostracan mitogenomes confirmed that J. (J.) hinumensis is most closely related to the family Micruropodidae, and supported the monophyly of the superfamily Gammaroidea.
Amphipods are a very important part of many freshwater and marine ecosystems (Barnard and Barnard Citation1983). Members of the superfamily Gammaroidea are widespread and play important functional roles in fresh and brackish water ecosystems in the northern hemisphere (Bousfield Citation1979; Jażdżewski Citation1980). The family Anisogammaridae Bousfield, Citation1979 is one of the Gammaroidean families endemic to the north Pacific Rim region (Tzvetkova Citation1975; Bousfield Citation1979,. The anisogammarid genus Jesogammarus Bousfield, Citation1979 consists of two subgenera (Annanogammarus Bousfield, Citation1979 and Jesogammarus Bousfield, Citation1979), which have been recorded in fresh and brackish waters in the Korean Peninsula, the Japanese archipelago, and the Chinese Continent (Bousfield Citation1979; Morino Citation1984; KS Lee and Seo Citation1990; Hou and Li Citation2004), and are currently known to include 20 species (Tomikawa et al. Citation2017). Of these, Jesogammarus (J.) hinumensis Morino, Citation1993 was originally described from a brackish lake in Japan, named Hinuma, and then on Jeju-island, Korea (Lee et al. Citation2019).
Previous studies have showed the phylogenetic position of Anisogammaridae within Gammaroidea (Tomikawa et al. Citation2010; Hou and Sket Citation2016). Hence, additional knowledge about the mitogenome of the anisogammarid species within Gammaroidea will improve our understanding of the phylogenetic relationships between gammaroidean amphipods.
Individuals of Jesogammarus were collected from brackish water in Korea (33°30.19′N, 126°53.51′E). Mitochondrial DNA extraction, sequencing, and gene annotation were performed following the methods described in Song et al. (Citation2016). The extracted mitochondrial DNA is maintained in the DNA collection at the National Institute of Biological Resources, Incheon, South Korea (deposit no. NIBRGR0000620078). A maximum-likelihood tree was constructed using IQ-tree 1.6.3 (Nguyen et al. Citation2015), based on the concatenated sequences of 10 PCGs (atp6, cox1, cox2, cox3, cytb, nad1, nad2, nad3, nad4, and nad5) from 25 eumalacostracan species, including the present sequence and two isopods as outgroup taxa (). The cox1 sequence of the extracted DNA was concordant with NCBI accession numbers LC052235 (Tomikawa Citation2015) and MN068364 (Lee et al. Citation2019), thus proving the taxonomic identity of the specimens under study.
Figure 1. Maximum-likelihood (ML) tree based on the mitogenome sequence of Jesogammarus (Jesogammarus) hinumensis (MT270126) and 25 other eumalacostracan species. The bootstrap supports are shown on each node.
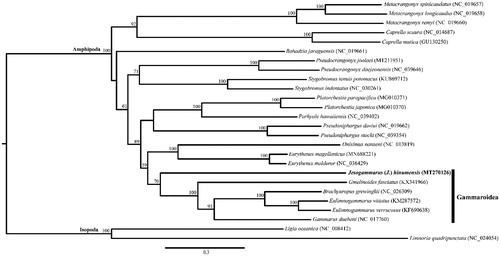
The complete mitogenome of J. (J.) hinumensis (GenBank accession no. MT270126) was 15,186 bp in length and contained the typical 13 PCGs, 22 tRNAs, 2 rRNAs, and a CR. The gene arrangement of J. (J.) hinumensis was concordant with the typical pan-crustacean ground pattern. The obtained maximum-likelihood tree supported the monophyly of the superfamily Gammaroidea, which includes J. (J.) hinumensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT270126.
Additional information
Funding
References
- Barnard JL, Barnard CM. 1983. Freshwater Amphipoda of the World, I. Evolutionary patterns and II. Handbook and bibliography. Mt. Vernon (VA): Hayfield Associates.
- Bousfield EL. 1979. The amphipod superfamily Gammaroidea in the northeastern Pacific region: systematics and distributional ecology. Bull Biol Soc Wash. 3:297–357.
- Hou ZE, Li S. 2004. Two new freshwater species of the genus Jesogammarus (Crustacea: Amphipoda: Anisogammaridae) from China. Raffles Bull Zool. 52:455–466.
- Hou ZE, Sket B. 2016. A review of Gammaridae (Crustacea: Amphipoda): the family extent, its evolutionary history, and taxonomic redefinition of genera. Zool J Linn Soc. 176(2):323–348.
- Jażdżewski K. 1980. Range extensions of some gammaridean species in European inland waters caused by human activity. Crustaceana. 6:84–107.
- Lee C-W, Tomikawa K, Min G-S. 2019. First record of the brackish water amphipod Jesogammarus (Jesogammarus) hinumensis (Amphipoda: Anisogammaridae) from Korea with DNA Barcode Analysis among Jesogammarus Species. Anim Syst Evol Divers. 35:151–155.
- Lee KS, Seo IS. 1990. One new species of freshwater Jesogammarus (Crustacea, Amphipoda, Anisogammaridae) from South Korea. Korean J Syst Zool. 6:251–260.
- Morino H. 1984. On a new freshwater species of Anisogammaridae (Gammaroidea: Amphipoda) from central Japan. Publ Itako Hydrobiol Stn. 1:17–23.
- Morino H. 1993. A new species of the genus Jesogammarus (Amphipoda: Anisogammaridae) from brackish waters of Japan. Publ Itako Hydrobiol Stn. 6:9–16.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Song J-H, Kim S, Shin S, Min G-S. 2016. The complete mitochondrial genome of the mysid shrimp, Neomysis japonica (Crustacea, Malacostraca, Mysida). Mitochondrial DNA A. 27(4):2781–2782.
- Tomikawa K. 2015. A new species of Jesogammarus from the Iki Island, Japan (Crustacea, Amphipoda, Anisogammaridae). ZK. 530:15–36.
- Tomikawa K, Kobayashi N, Mawatari SF. 2010. Phylogenetic relationships of superfamily Gammaroidea (Amphipoda) and its allies from Japan. Crust Res. 39(0):1–10.
- Tomikawa K, Nakano T, Hanzawa N. 2017. Two new species of Jesogammarus from Japan (Crustacea, Amphipoda, Anisogammaridae), with comments on the validity of the subgenera Jesogammarus and Annanogammarus. ZSE. 93(2):189–210.
- Tzvetkova NL. 1975. Littoral Gammaridae of northern and far eastern seas of the USSR. Leningrad: Academia Nauk USSR, Zoological Institute.
