Abstract
Terminalia neotaliala, a tropical tree species that originated from Madagascar, is a plant of high medical and horticultural value in Africa and China. This is the first study to present the complete chloroplast genome of T. neotaliala. The total chloroplast genome was 159,439bp in length, consisting of a large single copy (LSC) with 88,832bp, a small single copy (SSC) region of 19,013bp, and two inverted repeat (IRs) regions of 25,797bp. The overall GC content in the chloroplast genome was 36.9% and that of LSC, SSC and IRs were 34.7%, 30.5% and 43.0%, respectively. Of the 141 annotated genes, 96 were coding genes, eight were rRNA genes and 37 were tRNA genes. A phylogenetic analysis showed that in Myrtales, T. neotaliala is closely related to Laguncularia racemosa and Lumnitzera littorea.
Terminalia neotaliala is native to Madagascar. Like other Terminalia species, it is used in traditional medicine in Africa to treat skin and oral infections, and diseases such as diabetes (Tchuente Tchuenmogne et al. Citation2017; Toghueo et al. Citation2017). Terminalia neotaliala was introduced to China for horticultural purposes in the 1990s (Ou et al. Citation2013). It is widely used for landscaping in parks, campuses, streets, as well as in coastal areas due to its high tolerance to salinity (Huang et al. Citation2018). However, its natural populations are in decline due to agricultural expansion, logging and fire. Terminalia neotaliala is now listed as a ‘Vulnerable’ species in the IUCN Red List (Rakotoarisoa, Citation2019).
Here, we present the first complete chloroplast genome of T. neotaliala to provide genomic resources for identification and further investigation into its pharmacological properties.
Fresh healthy leaves of T. neotaliala were collected from a planted individual in the campus of Guangxi University, China (108.29460°E, 22.84251°N; China; voucher AKSW_TNGX01, deposited in the herbarium of the Ecological Genomics Team, Guangxi University, Nanning, China). The genomic DNA extraction, library construction and sequencing (on an Illumina HiSeq 2500 platform) were performed by Novogene (Beijing, China). The chloroplast genome was assembled de novo using NOVOPlasty (Dierckxsens et al. Citation2017), and annotated using GeSeq (Tillich et al. Citation2017) and Geneious v11.0.4 (https://www.geneious.com, Kearse et al. Citation2012).
The complete chloroplast genome of T. neotaliala (Genbank accession: MN251603) is a double-stranded circular DNA. It is 159,439 bp in length, consisting of a large single copy (LSC) with 88,832 bp, a small single copy (SSC) region of 19,013 bp, and two inverted repeat (IRs) regions of 25,797 bp. The overall GC content in the chloroplast genome was 36.9% and that of LSC, SSC and IRs were 34.7%, 30.5% and 43.0%, respectively. Of the 141 annotated genes, 96 were coding genes, eight were rRNA genes and 37 were tRNA genes.
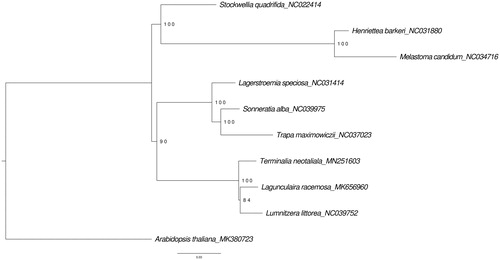
A phylogeny of Myrtales was constructed using eight complete chloroplast genome sequences of species in Myrtales obtained from GenBank, and chloroplast genomes of T. neotaliala (Genbank accession: MN251603) and Laguncularia racemosa (Genbank accession: MK656960) assembled in this study. The phylogeny was constructed by performing a maximum likelihood analysis using RAxML-ng 0.9.0 (Kozlov et al. Citation2019) with 1000 bootstrap repeats, after determining the best fitting model by ModelTest-ng (Diego et al. Citation2019). The phylogenetic tree () showed that T. neotaliala is closely related to L. racemosa and Lumnitzera littorea within the Myrtales.
Figure 1. Maximum likelyhood (ML) phylogenetic tree constructed using 10 complete chloroplast genome of Myrtales rooted with Arabidopsis thaliana (Brassicales). The numbers on nodes are bootstrap support values.
This is the first study to present the complete chloroplast genome of T. neotaliala. This chloroplast genome can be used in phylogenetic studies to assess the evolution of medical properties in Terminalia (e.g. Tchuente Tchuenmogne et al. Citation2017) and to date the evolutionary divergence within Myrtales (e.g. Berger et al. Citation2016).
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov, GenBank accession number MN251603, MK656960.
Additional information
Funding
References
- Berger BA, Kriebel R, Spalink D, Sytsma KJ. 2016. Divergence times, historical biogeography, and shifts in speciation rates of Myrtales. Mol Phylogenet Evol. 95:116–136.
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. 2019. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Huang XL, Lin FF, Zhang MY, Zhu LQ, Zhao LJ. 2018. Effects of salt stress on growth and osmoregulatory substances in Terminalia neotaliala Capuron seedlings. J Southern Agric. 49(7):1364–1369.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Ou B, Lin XJ, Chen LH, Tan CQ, Wang RC. 2013. Effects of different coverage on seedlings emergence of Terminalia mantaly H. Perrier. Trop Forestry. 41(3):43–45.
- Rakotoarisoa SE. 2019. Terminalia neotaliala. The IUCN Red List of Threatened Species 2019. [accessed 2020 May 3]. https://dx.doi.org/10.2305/IUCN.UK.2019-2.RLTS.T128091759A128098510.en.
- Tchuente Tchuenmogne MA, Kammalac TN, Gohlke S, Kouipou RMT, Aslan A, Kuzu M, Comakli V, Demirdag R, Ngouela SA, Tsamo E, et al. 2017. Compounds from Terminalia mantaly L. (Combretaceae) stem bark exhibit potent inhibition against some pathogenic yeasts and enzymes of metabolic significance. Medicines. 4(1):6.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Toghueo RMK, Zabalgogeazcoa I, Vazquez de Aldana BR, Boyom FF. 2017. Enzymatic activity of endophytic fungi from the medicinal plants Terminalia catappa, Terminalia mantaly and Cananga odorata. S Afr J Bot. 109:146–153.
