Abstract
We sequenced and assembled the complete mitochondrial genome sequence of the Meretrix lusoria, from Kumamoto, Japan. The length of mitogenome is 20,180 bp, including 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes. The nucleotide composition of the mitogenome was 25.73% for A, 42.41% for T, 9.35% for C, and 22.49% for G. The AT and GC skewness of mitogenome sequence are −0.245 and 0.412, showing the T-skew and G-skew. The reconstructed phylogenetic relationships of 25 Bivalvia species based on 12 protein-coding genes were highly supported and the clade of all Meretrix clams included had a support value of 99%. Our results shall provide a better understanding in the evolutionary histories of the Veneroida and relative species.
The Meretrix clams are widely distributed along the coast of East Asia to Southeast Asia. Three Meretrix species, Meretrix lusoria, M. lamarckii, and Meretrix sp. are distributed in Japan. The M. lusoria and M. lamarckii can be found in the shell mounds of the Jomon period about 8000 years ago (Sakatsume Citation1959). According to the survey on marine fishery production, M. lusoria was an important commercial bivalve in Japan, particularly in Tokyo Bay, Ise Bay, and the Ariake Sea (Henmi et al. Citation2014). Although the abundances of the clams have decreased greatly in many regions, the Shirakawa river and Midorikawa river are still the important habitats for the natural population (Nakamura Citation2013; Hashiguchi et al. Citation2014). However, it is difficult to identify the hard calms only depending on the morphological characteristics. The DNA molecular information is useful and easy to identify them correctly and quickly, especially the sequence of mitochondrial DNA.
In this study, the Asia hard clam was collected from Shirakawa river (32°46′36″N; 130°36′42″E) in Kumamoto, Japan and stored in Fisheries Research Institute in Keelung, Taiwan with accession number FIRM10017. The total of 4.3 Gb next-generation sequencing paired-end reads was used to assemble the complete mitogenome sequence of the M. lusoria. The CLC Genomics Workbench (QIAGEN) was used for sequencing reads quality analysis, reads trimming, and de novo assembling. The locations of the protein-coding genes, ribosomal RNAs (rRNAs), and transfer RNAs (tRNAs) were predicted by using MITOS Web Server (Bernt et al. Citation2013) and identified by alignment with other mitogenome sequence of Meretrix shellfish. The AT and GC skew was calculated according to the following formulas: AT skew = (A-T)/(A + T) and GC skew = (G-C)/(G + C) (Perna and Kocher Citation1995).
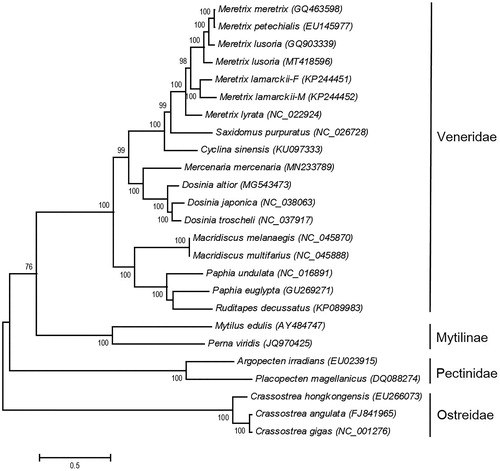
The complete mitogenome of M. lusoria from Kumamoto is 20,180 bp in length (GenBank Accession No.MT418596), including 13 protein-coding genes, two rRNA genes, and 22 tRNA genes. The nucleotide composition of the mitogenome was 25.73% for A, 42.41% for T, 9.35% for C, and 22.49% for G. The AT content was 68.15% and similar to congeneric species. The AT and GC skewness of mitogenome sequence were −0.245 and 0.412, showing the T-skew and G-skew. The skew statistics (AT and GC) of protein-coding gene were −0.341 and 0.392, showing the same T-skew and G-skew. The mitogenome sequence identity of 13 protein-coding genes is 79.17–88.59% with other Meretrix clams. We reconstructed the phylogenetic relationships of Veneroida and relative species based on 12 protein-coding genes (excluding the atp8 gene) of 25 mitogenomes with Maximum-Likelihood (ML) criteria. Bootstrap values (1000 replications) greater than 70% are shown at the branch nodes (). The majority of nodes had support values higher than 70% and 17 were up to 100% supported. The Meretrix clams were grouped in the same clade and the clade of Veneroida was highly supported. The phylogenetic position of M. lusoria (MT418596) is sister to M. meretrix (GQ463598), M. petechialis (EU145977), and M. lusoria (GQ903339). Hsiao et al. (Citation2019) observed that the cox1 gene sequence of GQ903339 was grouped with the Meretrix sp. nov., a new species in Taiwan and southern China. Our results shall provide a better understanding in the evolutionary histories of the Veneroida and relative species. In this study, mitogenomic sequence data will provide useful information for further studies for the population genetics, speciation, biogeography, and conservation of M. lusoria in the future.
Figure 1. Phylogenetic tree of the 25 Bivalvia species based on the sequence of 12 protein-coding genes. The tree was reconstructed with the Maximum-Likelihood (ML) criteria using MEGA v.6 (Tamura et al. Citation2013). Bootstrap values (1000 replications) greater than 70% are shown at the branch nodes.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in nucleotide database of NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov, accession number MT418596.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Hashiguchi M, Yamaguchi J, Henmi Y. 2014. Distribution and movement between habitats with growth of the hard clam Meretrix lusoria in the Shirakawa–Midorikawa estuary of the Ariake Sea. Fish Sci. 80(4):687–693.
- Henmi Y, Kobayashi S, Yamaguchi J, Hashiguchi M. 2014. Recruitment and movement of the hard clam Meretrix lusoria in a tidal river of northern Kyushu. Fish Sci. 80(4):705–714.
- Hsiao ST, Chuang SC, Wu YH, Chang JW, Chin CP, Yeh HM, Henmi Y, Chen JR. 2019. Preliminary studies for the habitat conditions of the Asian hard clam Meretrix spp. nov. in Tamsui estuary, Taiwan. Proceedings of the 2019 CWMD International Conference 2019, September 19–21, 2019, Kumamoto University, Japan. p. 474–482.
- Nakamura Y. 2013. Secretion of a mucous cord for drifting by the clam Meretrix lusoria (Veneridae). Plankton Benthos Res. 8(1):31–37.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41(3):353–358.
- Sakatsume N. 1959. Nihon Kaizuka Chimei-hyo. (A list of shell middens in Japan). Tokyo, Japan: Doyokai (in Japanese).
- Tamura k, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
