Abstract
The complete mitochondrial genome of the highly venomous blue-lined octopus, Hapalochlaena fasciata (Hoyle, 1886), was analyzed by the primer walking method. Its mitogenome was 15,479 bp in total length, comprising 13 protein-coding genes, 2 ribosomal RNA genes, and 23 transfer RNA genes. In the phylogenetic tree, the gene content and order were congruent with those of typical octopodiform species. The mitogenomic sequence presented could be very useful as the first record of the complete mitogenome for the genus Hapalochlaena.
The genus Hapalochlaena are only octopuses known to be capable of causing toxic morbidity or mortality in humans (White and Meier Citation1995). Among them, the shallow-water benthic octopus Hapalochlaena fasciata, which is also called as the blue-lined octopus, is most commonly found around intertidal rocky shores and coastal waters between Australia and the Pacific Ocean north to Japan (Williamson et al. Citation1996; Kohtsuka Citation2006; Kim et al. Citation2012). In Korea, the appearance of H. fasciata is increasing every year (Kim et al. Citation2018), but there are very little scientific records. In this study, we reported the complete mitochondrial genome (mitogenome) of the highly venomous blue-lined octopus, H. fasciata from the southern coastal waters, Korea.
A specimen of H. fasciata was collected in the southern coastal water of Korea, at 15 Nov. 2019 (34°26′17.93ʺN, 127°51′32.36ʺE) and deposited at the National Marine Biodiversity Institute of Korea (voucher no. MABIK MO00176321) after ethanol fixation. Its genomic DNA was extracted from a tissue around the mouth with DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). Its complete mitogenome was amplified through two independent and overlapping PCR runs, and the PCR products were sequenced using 25 sequencing primers. The complete mitogenomic sequence was deposited in the GenBank under the accession number MT497543.
All mitogenomic sequences of the species belonging to the superorder Octopodiformes were retrieved from GenBank in NCBI (http://www.ncbi.nlm.nih.gov/). They were aligned together with that of H. fasciata in this study and refined manually. For the phylogenetic analysis, the nucleotide matrix of 13 protein-coding genes was created and divided into three partitions, consisting of 3706 bp each for the first, second, and third bases of each codon triplet, respectively. A phylogenetic tree was reconstructed using RAxML 7.0.4 (Stamatakis Citation2006) for maximum-likelihood (ML) analysis.
The complete mitogenome of H. fasciata was a circular molecule of 15,479 bp in total length, consisting of 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes, of which gene content is identical those of typical octopuses. Its gene order was also identical to those of typical octopuses, but different from those of decapodiform species.
With the mitogenomic sequence of H. fasciata, a phylogenetic tree was reconstructed by the ML method, using the nucleotide sequence matrix from 13 protein-coding genes (). In the resulting tree, octopodiform species formed the monophyletic group with 100% bootstrap value with respect to the two decapodiform outgroups. Among the lineage, H. fasciata in this study consistently emerged among Amphioctopus species with close phylogenetic relationship to Amphioctopus aegina, A. marginatus, and A. neglectus with 85% bootstrap value.
Figure 1. Maximum-likelihood (ML) phylogeny based on the complete mitochondrial genomes from the octopuses belonging to the superorder Octopodiformes. The nucleotide sequence matrix included the three codon positions of the 13 protein-coding genes. A bootstrap value above 50% in the ML analysis is indicated at each node. Hapalochlaena fasciata analyzed in this study is shown in bold.
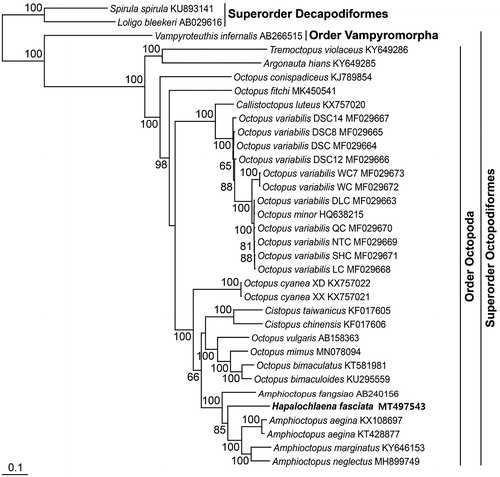
In this study, the complete mitogenomic sequence of the genus Hapalochlaena was analyzed for the first time, and the phylogenetic tree among octopuses was reconstructed with reference to their mitogenomic sequence data. Our data will provide baseline data for the molecular identification, geographical distribution and dispersal, population genetics, and early warning for the highly venomous marine cephalopod species, H. fasciata in the Korean coastal waters.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT497543.
Additional information
Funding
References
- Kim HS, Kwun HJ, Bae H, Park J. 2018. First reliable record of the blue-lined octopus, Hapalochlaena fasciata (Hoyle, 1886) (Cephalopoda: Octopodidae), from Jeju Island, Korea. J Asia-Pac Biodivers. 11(1):21–24.
- Kim JH, Suzuki T, Shim KB, Oh EG. 2012. The widespread distribution of the venomous and poisonous blue-lined octopus Hapalochlaena spp., in the East/Japan Sea: possible effects of sea warming. Fish Aquat Sci. 15(1):1–8.
- Kohtsuka H. 2006. Record of blue-ringed octopus Hapalochlaena cf. fasciata (Mollusca: Cephalopoda) from Oki Island, Sea of Japan. Bull Hoshizaki Green Found. 9:226.
- White J, Meier J. 1995. Handbook of clinical toxicology of animal venoms and poisons. Vol. 236. New York, NY: CRC Press.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Williamson JA, Fenner PJ, Burnett JW, Rifkin JF. 1996. Venomous and poisonous marine animals: a medical and biological handbook. Sydney, Australia: UNSW Press.
