Abstract
Geodorum eulophioides Schltr., is a critically Endangered orchid (IUCN). In this study, we report the first complete chloroplast (cp) genome of G. eulophioides to provide the underlying information for genetic breeding and conservation studies of this species. The cp genome sequence of G. eulophioides is 149,466 bp in length, which contains one large single-copy region (LSC, 85,436 bp), one small single-copy region (SSC, 14,086 bp), and two inverted repeat regions (IRs, 24,972 bp). The cp genome encoded 177 genes, of which 106 were unique genes (78 protein-coding genes, 24 tRNAs, and 4 rRNAs). Phylogenetic analysis showed that G. eulophioides is closely related to the genera Eulophia.
Geodorum Jacks (Orchidaceae) has approximately 10 species that are distributed from tropical Asia to Australia and the South-West Pacific Islands. There are six species in China, including two endemic species (Chen et al. Citation2009).
Geodorum eulophioides Schltr. (1921) is a critically Endangered orchid (IUCN), which grows along the valley with an elevation of 800 meters. It has a few known populations: two found in southwest China (Liu Citation2010) and a location in central Myanmar (Tanaka et al. Citation2011). In this study, we sequenced the complete plastome of G. eulophioides to provide the underlying information for genetic breeding and conservation studies of this species.
Leaf samples of G. eulophioides were obtained from the Orchid Conservation and Research Center of Shenzhen and specimens that were deposited in the China National Orchid Conservation Center Herbarium (NOCC; specimen code Z.J. Liu 8824). Total genomic DNA was extracted from fresh leaves using the modified CTAB procedure method (Doyle and Doyle Citation1987) and sequenced by using Illumina Hiseq 4000 platform (San Diego, CA, USA). After assembled by MITObim v1.8 (Hahn et al. Citation2013), the obtained scaffolds and contigs were annotated with CpGAVAS (Liu et al. Citation2012), finally adjusted by Geneious version 11.1.15 (Kearse et al. Citation2012). This newly obtained complete cp genome sequence was submitted to GenBank (ID:MK848065).
The cp genome sequence of G. eulophioides is 149,466 bp in length, which contains one large single-copy region (LSC, 85,436 bp), one small single-copy region (SSC, 14,086 bp), and two inverted repeat regions (IRs, 24,972 bp). The cp genome encoded 177 genes, of which 106 were unique genes (78 protein-coding genes, 24 tRNAs, and 4 rRNAs).
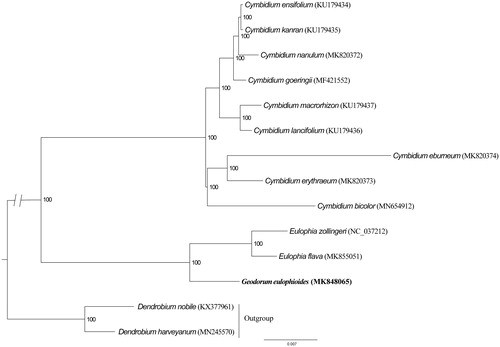
To further investigate its phylogenetic position, we used RAxML-HPC2 on XSEDE 8.2.12 (Stamatakis et al. Citation2008) to construct a maximum likelihood tree with other 13 published cp genomes of Orchidaceae. We computed the branch support with 1000 bootstrap replicates by Stamatakis et al. (Citation2008). Phylogenetic analysis showed that G. eulophioides is closely related to the genera Eulophia (). The determination of the complete plastid genome sequences provided new molecular data, which is helpful to illuminate evolution mechanism in Orchidaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MK848065].
Additional information
Funding
References
- Chen XQ, Cribb PJ, Gale SW. 2009. Orchidaceae. In: Wu ZY, Raven P, Hong DY, editors. Flora of China. Vol. 25. Beijing, China and St. Louis, Missouri, USA: Science Press and Missouri Botanical Garden Press. 25:258–260.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Liu H. 2010. Geodorum eulophioides: challenges for conserving one of the rarest Chinese orchids. Orchids. 79(3):161–162.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57(5):758–771.
- Tanaka N, Yukawa T, Htwe KM, Koyama T, Murata J. 2011. New or noteworthy plant collections from Myanmar (7): fourteen additional species of Orchidaceae. Acta Phytotaxonomica et Geobotanica. 61:161–165.