Abstract
Ostericum citriodorum is a traditional Chinese medicinal herb endemic to Southeast and South China, but now is becoming very rare because of rapid habit loss. The complete chloroplast genome of O. citriodorum was sequenced herein and suggested that the complete chloroplast genome was 155,919 bp in length, comprising the large single-copy (LSC) region of 85,393 bp, the small single-copy (SSC) region of 19,760 bp, and a pair of inverted regions (IRs) of 25,383 bp. Totally 127 genes were distributed in the whole genome, including 4 rRNAs, 37 tRNAs, and 81 protein coding genes. The G + C content of this chloroplast genome was 38%. Phylogenetic inference revealed that O. citriodorum was accompanied with Pterygopleurum neurophyllum and sister to O. palustre, indicating a close relationship between Ostericum and Pterygopleurum.
Ostericum citriodorum (Hance) R.H. Shan & C.Q. Yuan is a striking perennial umbelliferous herb that has been used as a traditional Chinese medicine for treating angina pectoris (Shan Citation1992; Luo et al. Citation2020). As an endemic plant to China, this species is widespread in the hills or low mountains of Southeast and South China, but now is seriously threatened by rapid habit loss (Sheh et al. Citation2005; Liao et al. Citation2013). This research generated the first complete chloroplast genome of O. citriodorum aim to provide genomic information for the effective conservation and accurate identification of this species.
Fresh materials were obtained from the cultivated O. citriodorum in the green house of Sichuan University, Chengdu, China, which was transplanted from Luzhai, Guangxi, China. Both the voucher specimens and DNA samples were deposited in the herbarium of Sichuan University (accession no. lcy20120712). Morphological analysis was conducted by Karyotype (Altınordu et al. Citation2016). Total genomic DNA was extracted using Plant Genomic DNA Kit. The isolated genomic DNA was constructed to an average 400 bp paired-end (PE) library using the Illumina Hiseq platform (Illumina, San Diego, CA), and sequenced using Illumina genome analyzer (Hiseq PE150). The gene reconstruction, annotation, and analysis were carried out in Geneious Prime 2020.1.2 (Kearse et al. Citation2012), referring to Pterygopleurum neurophyllum (Maxim.) Kitag. (GenBank no. NC033345.1). The complete assemble genome sequence was deposited in the GenBank (accession no. MT501096).
The complete chloroplast genome of O. citriodorum was totally 155,919 bp in length with a circular form, and divided into four regions: the large single-copy region (LSC) of 85,393 bp, the small single-copy (SSC) region of 19,760 bp, and two inverted regions (IRs) of 25,383 bp each. The total GC content of this species was 38%, including 36% for the LSC, 31% for the SSC, and 43% for each IR. There were four rRNAs, 37 tRNAs, and 81 protein coding genes, for a total of 127 genes was annotated through the whole chloroplast genome. The tRNA coding genes were widely distributed in the genome, including 22 located in the LSC, 7 in each IR and only 1 in the SSC, while the rRNA coding genes exclusively placed in the IRs.
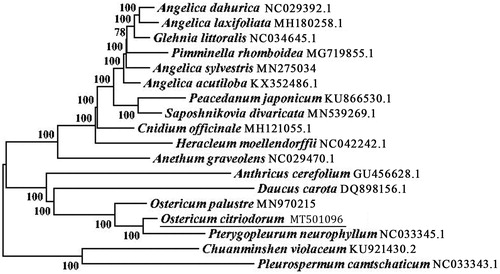
An alignment comprising the complete chloroplast genome sequences of O. citriodorum and other 17 related taxa of Apiaceae was performed using MAFFT (Katoh et al. Citation2002), and then trimmed and manually corrected using trimAl v1.4 (Capella-Gutierrez et al. Citation2009). Afterwards, neighbour-joining (NJ) trees were generated using MEGA7.0 (Kumar et al. Citation2016) with 1000 bootstrap replicates (). The result suggested that O. citriodorum was accompanied with Pterygopleurum neurophyllum and sister to O. palustre, which was consistent with our previous research on morphology and molecular phylogeny of Ostericum (Liao et al. Citation2013; Li et al. Citation2017; Zhang et al. Citation2018; Liao et al. Citation2020), and also indicated a new point of view on the relationship between the genera of Ostericum and Pterygopleurum.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All the chloroplast genome sequences used in this study are derived from the GenBank database at http://www.ncbi.nlm.nih.gov/genbank, and the reference numbers are DQ898156.1, GU456628.1, KU866530.1, KU921430.2, KX352486.1, MG719855.1, MH121055.1, MH180258.1, MN275034, MN539269.1, MN970215, MT501096, NC029392.1, NC029470.1, NC033343.1, NC033345.1, NC034645.1, NC042242.1, respectively.
Additional information
Funding
References
- Altınordu F, Peruzzi L, Yu Y, He XJ. 2016. A tool for the analysis of chromosomes: KaryoType. Taxon. 65(3):586–592.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li ML, Liao CY, Ye C, Zhang JZ, Feng TY, Zhou B. 2017. Analysis of leaf epidermal micromorphological characters of Ostericum Hoffm. (Apiaceae). Acta Bot Boreal-Occident Sin. 37 (8):1540–1549.
- Liao CY, Chen XF, Tan JB, Gao Q. 2020. The complete chloroplast genome of Ostericum palustre (Apiaceae). Mitochondr DNA B. 5 (2):1357–1358.
- Liao CY, Downie SR, Li QQ, Yu Y, He XJ, Zhou B. 2013. New insights into the phylogeny of Angelica and its Allies (Apiaceae) with emphasis on East Asian species, inferred from nrDNA, cpDNA, and morphological evidence. System Bot. 38(1):266–281.
- Luo TS, Chen ZW, Wang FY, Yin SS, Liu P, Zhang J, Yang ZH. 2020. Endothelium-independent vasodilatory effects of isodillapiolglycol isolated from Ostericum citriodorum. Molecules. 25(4):885.
- Shan RH. 1992. Umbelliferae. In: Shan RH, Sheh ML, editors. Flora reipublicae popularis sinicae, vol. 55. Beijing: Academia Sinica; p. 13–62.
- Sheh ML, Pu FT, Pan ZH, Watson MF, Cannon JFM, Holmes-Smith I, Kljuykov EV, Phillippe LR, Pimenov MG. 2005. Apiaceae. In Z. Y. Wu and P. H. Raven. St. Louis, Missouri: Missouri Botanical Garden Press and Beijing: Science Press; p. 1–205.
- Zhang SY, Liao CY, Li ML, Chen Y, Zhou B. 2018. Research on pollen morphologies of Ostericum Hoffm. (Apiaceae) of eight species from seventeen populations. Acta Bot Boreal Occident Sin. 38(12):2224–2235.