Abstract
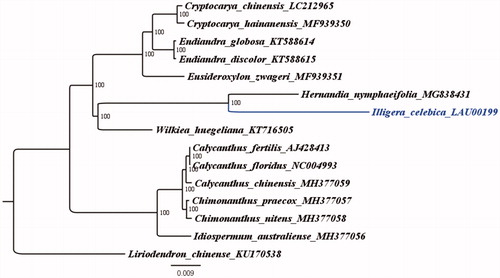
Illigera celebica is an evergreen woody vine that belongs to genus Illigera Bl in the family Hernandiaceae and has medicinal value. The complete chloroplast genome of I. celebica was sequenced to determine its phylogenetic location with respect to the other species under the Laurales. Its whole chloroplast genome is 156,123 bp in length, and comprises a large single-copy region (LSC, 84,913 bp), a small single-copy region (SSC,18,775 bp), and a pair of inverted repeats (IRs, 26,217 bp). The overall GC content is 39.2% (LSC, 37.8%; SSC, 33.9%; IR, 43.4%). Maximum likelihood phylogenetic analysise (TVM + F + R2 model) was conducted using 15 complete chloroplast genomes of Laurales, and the results confirmed that Hernandia nymphaeifolia and Wilkiea huegeliana were located in the same lineage.
Illigera celebica is a kind of evergreen woody vine that belongs to genus Illigera Bl in the family Hernandiaceae. I. celebica is widely distributed in Yunnan, Guangxi, and Guangdong provinces (Chinese Flora Editorial Board, Chinese Academy of Sciences Citation1982) of south China, and in other countries, such as Vietnam, Thailand, and Cambodia (http://foc.iplant.cn/). The root and stem of I. celebica can dispel wind, dehumidify, and relieve pain (Huang Citation1985; Gao Citation2007). At present, genus Illigera has no clear phylogenetic system. Therefore, the complete chloroplast genome of I. celebica was obtained by high-throughput sequencing to reconstruct a phylogenetic tree to better understand the relationships of I. celebica and other Laurales species.
The healthy young leaves of I. celebica were freshly picked from Xishuangbanna Tropical Botanical Garden (XTBG) in Yunnan, China (101.2713°E longitude, 21.9170°N latitude; 540 m). DNA was extracted using modified CTBA method (Cai et al. Citation2014) and the specimens were stored in XTBG’s Biodiversity Research Group (Registry No. SWFU-SY36764). The whole chloroplast genome was sequenced according to the method of Yang et al. (Citation2014). The whole nine pairs of universal primers were sequenced by remote polymerase chain reaction for next-generation sequencing. The publicly available chloroplast genome of Eusideroxylon zwageri (Accession No.MF939351) was used as reference. The chloroplast genome of I. celebica was assembled using the GetOrganelle software (Jin et al. Citation2018) and annotated through the Geneious 8.1.3 software (Biomatters Ltd., Auckland, New Zealand).
The chloroplast genome of I. celebica (LAU00199) with a length of 156,123bp, which is 1,639 and 1,454 bp smaller than those of Hernandia nymphaeifolia (157,762 bp, MG838431) and E. zwageri (157,577 bp, MF939351). It was also 22,963 bp large than that of Wilkiea huegeliana (133,160 bp, KT716505). The results showed that the complete genome of I. celebica is composed of a large single-copy region (LSC, 84,913 bp), a small single-copy region (SSC, 18,775 bp), and a pair of inverted repeats (IRs, 26,217 bp). The overall GC content is 39.2% (LSC, 37.8%; SSC, 33.9%; IR, 43.4%). The chloroplast genome of I. celebica contains 112 unique genes, which are composed of 76 protein-coding genes, 8 are rRNA genes, and 37 tRNA genes.
The evolutionary relationship between I. celebica and other Laurales species was determined based on the complete sequence of the chloroplast genome of I. celebica and the reconstruction of a phylogenetic tree from the thirteen published chloroplast genomes of family Lauraceae (). In addition, Liriodendron chinense (Accession Number: KU170538) was treated as an outgroup. A maximum-likelihood (ML) analysis based on the TVM + F + R2 model was performed with iqtree version 1.6.7 program using 1000 bootstrap replicates (Nguyen et al. Citation2015). The ML phylogenetic tree of Liriodendron chinense showed 100% bootstrap values at each node, confirmed that H. nymphaeifolia and Wilkiea huegeliana belong to the same lineage (Song et al. Citation2019).
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The chloroplast data of the I. celebica will be submitted to Laurales Chloroplast Genome Database (https://lcgdb.wordpress.com). Accession numbers are LAU00199.
Additional information
Funding
References
- Cai WJ, Xu DB, Lan X, Xie HH, Wei JG. 2014. A new method for the extraction of fungal genomic DNA. Agric Res Appl. 152:1–5.
- Chinese Flora Editorial Board, Chinese Academy of Sciences. 1982. Flora of China. Vol. 31. J B Sci Press(China); p. 476.
- Gao Z. 2007. It is made up of plant extracts such as Sinomenium spp., Aconitum carmichaeli Debx. and Paeonia lactiflora Pall to treat arthritis and pain. Drugs Clinic. 22(4):181–182.
- Huang X. 1985. A new species and medicinal plant of the same genus from Illigera BI of Guangxi. Guihaia. 5(1):17–20.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assemble of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479. doi:10.1101/256479.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Song Y, Yu WB, Tan YH, Jin JJ, Wang B, Yang JB, Liu B, Corlett RT. 2019. Plastid phylogenmics improve phylogenetic resolution in the Lauraceae. J Sytemat Evol. doi:10.1111/jse.12536.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.