Abstract
The complete mitochondrial genome (mitogenome) of Lymantria sugii (Diptera: Tephritidae: Dacinae) was sequenced and annotated. The mitochondrial genome is 15,614 bp (GenBank No. MT265380), containing 80.4% A + T (A 39.1%, C 7.3%, G 12.2%, and T 41.3%), that is heavily biased toward A and T nucleotides. All PCGs were started with ATN (ATA/ATG/ATT/ATC) and were terminated with TAR (TAA/TAG) excepting ND4, which ended with single T. Additionally, the phylogenetic tree confirmed that L. sugii clustered with L. umbrosa, L. dispar and Lymantria sp. The current study would be enrich the mitogenomes of the Lymantriinae.
Lymantria sugii belongs to Lymantria genus. Many species in Lymantria genus are known as significant forestry pests. Currently, the Lymantria genus includes 173 species, containing 12 confirmed subgenera all over the world. Among them, 31 species of 3 subgenera were selectively reviewed as potential invaders to North America (Kang et al. Citation2015). The identification of Lymantria sugii is mainly based on morphology and partial sequence. Presently, we have sequenced and determined the complete mitochondrial genome (mitogenome) using next-generation sequencing method for the first time, which might facilitate future studies on the Lymantriinae.
The genome DNA was extracted from male adult of Lymantria sugii which was collected in Zhuanghe area, Dalian city, China (E 123°10′43.79″, N 39°42′14.34″), in September 2019, and the voucher specimen are deposited in the Insect Collection of Zhejiang Academy of Science & Technology for Inspection & Quarantine (ZAIQ) with label number ZAIQ-LL-SM-1901. These sequences were assembled using Geneious Primer (Kearse et al. Citation2012), version 10.2.3 (http://www.geneious.com/). Additionally, all tRNAs were found by MITOS server (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013) and tRNA scan-SE server (Lowe and Chan Citation2016) for annotation. The neighbor-joining (NJ) tree was constructed to investigate the molecular taxonomic position of Lymantria sugii basing on nucleotide sequences of 13 protein-coding genes and 2 rRNA genes using MEGA 6.0 (Tamura et al. Citation2013) from alignments created by the MAFFT (Katoh and Standley Citation2013).
The complete mitogenome of Lymantria sugii is 15,614 bp (GenBank No. MT265380), containing 22 transfer RNA genes (tRNAs, 1470 bp), 13 protein-coding genes (PCGs, 11,074 bp), 2 ribosomal RNA genes (rRNAs, 2181 bp), and one non-coding region (Control region, 391 bp). The whole genome contained 39.1% A, 7.3% C, 12.2% G, and 41.3%T, showing an obvious A + T bias (80.4%). The AT-skew and GC-skew for the whole mitogenome is –0.0274 and 0.2513 respectively. All PCGs were started with ATN (ATA/ATG/ATT/ATC) and terminated with TAR (TAA/TAG) excepting ND4, which ends with single T.
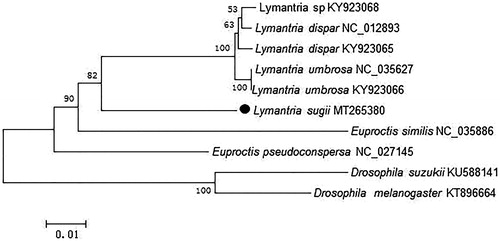
The phylogenetic relationships of L. sugii were reconstructed using the neighbor-joining (NJ) method with 1000 bootstrap replicates basing on concatenated nucleotides of the 13 PCGs and 2 rRNAs with 13,194 bp, Dosophila suzukii and Drosophila melanogaster were used as outgroup (). The phylogenetic tree confirmed that L. sugii clustered with L. dispar, Lymantria sp. and L. umbrosa. Presently, the studies recording L. sugii were limited, and we believe that our data could be useful for further study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number MT265380.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Kang TH, Lee KS, Lee HS. 2015. DNA barcoding of Korean Lymantria Hübner, 1819 (Lepidoptera: Erebidae: Lymantriinae) for quarantine inspection. Commod Treat Quarant Entomol. 108(4):1597–1611.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.