Abstract
Rumex nepalensis (Polygonaceae) is a fairly common perennial herb of high altitudes. In this study, we determined the complete chloroplast genome (plastome) of R. nepalensis with genome-skimming method. The complete plastome of R. nepalensis was 159,110 bp in length with a quadripartite structure, including a large single-copy region of 84,810 bp, a small single-copy region of 13,044 bp, and a pair of inverted repeats regions of 30,628 bp. The overall guanine-cytosine (GC) content was 37.5%. A total of 112 unique genes was annotated in this plastome, including 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. In the ML tree, R. nepalensis was sister to R. crispus, and Rumex was sister to a clade comprising Rheum and Oxyria within Polygonaceae famliy.
Keywords:
Rumex nepalensis (Polygonaceae) is a fairly common perennial herb of high altitudes (Farooq et al. Citation2013). It grows between 800–4000 m in Afghanistan, China, India, Indonesia, Japan, Myanmar, Nigeria, Nepal, Pakistan, Tajikistan, Vietnam, South-west Asia, Turkey, Bhutan and South Africa (Kumar et al. Citation2018). For thousands of years, R. nepalensis has been served as the basis of traditional medicine systems in Nigeria, India, China and Indonesia (Ahmad et al. Citation2012; Yi et al. Citation2012; Kumar et al. Citation2018). It can also be used as vegetable, forage, coloring agent, and flavoring (Venkataramegowda Citation2012; Kumar et al. Citation2018). In this study, we reported the plastome of R. nepalensis for resolving its phylogenetic position.
Fresh leaves of R. nepalensis were sampled from Kunming Botanic Garden, Chinese Academy of Sciences (Yunnan, China; 25°8′N, 102°44′E). Voucher specimen (SD251) was deposited at College of Life Sciences, Shandong Normal University. Total genomic DNA was extracted using the modified CTAB method as described in Qu, Fan, et al. (Citation2019). Total genomic DNA was then sequenced with the Novaseq platform. Plastome assembly was conducted with Organelle Genome Assembler (OGA) (Qu, Fan, et al. Citation2019). Plastome annotation was conducted with Plastid Genome Annotator (PGA) (Qu, Moore, et al. Citation2019), by manual correction with Geneious v9.1.4. The complete plastome was submitted to GenBank with accession number MT457825.
The complete plastome of R. nepalensis was 159,110 bp in length with a quadripartite structure, including a large single-copy region of 84,810 bp, a small single-copy region of 13,044 bp, and a pair of inverted repeats regions of 30,628 bp. The overall guanine-cytosine (GC) content was 37.5%. A total of 112 unique genes was annotated in this plastome, including 78 protein-coding genes, 30 tRNA genes, and four rRNA genes. Among them, ten protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12, and rps16) and six tRNA genes (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained one intron, and two protein-coding genes (clpP and ycf3) contained two introns.
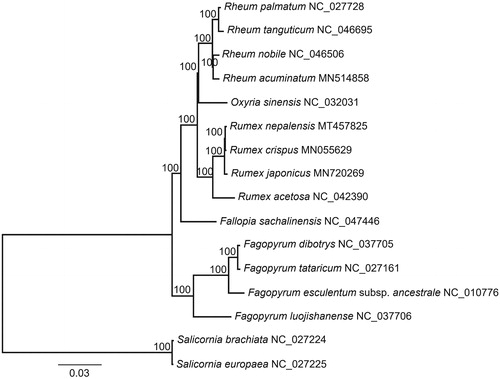
To determine the phylogenetic position of R. nepalensis, a maximum likelihood tree of 14 Polygonaceae species () was reconstructed with RAxML v8.2.10 (Stamatakis Citation2014), based on alignment of all shared genes with MAFFT v7.313 (Katoh and Standley Citation2013). In the ML tree, R. nepalensis was sister to R. crispus, and Rumex was sister to a clade comprising Rheum and Oxyria within Polygonaceae famliy.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT457825.
Additional information
Funding
References
- Ahmad K, Kayani W, Hameed M, Ahmad F, Nawaz T. 2012. Floristic diversity and ethnobotany of senhsa, district kotli, Azad Jammu & Kashmir (Pakistan). Pakistan J Bot. 44:195–201.
- Farooq U, Pandith SA, Singh Saggoo MI, Lattoo SK. 2013. Altitudinal variability in anthraquinone constituents from novel cytotypes of Rumex nepalensis Spreng—a high value medicinal herb of North Western Himalayas. Ind Crop Prod. 50:112–117.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar V, Shaikh S, Barve P, Shriram V. 2018. A critical review on Nepal Dock (Rumex nepalensis): a tropical herb with immense medicinal importance. Asian Pac J Trop Med. 11:405.
- Qu XJ, Fan SJ, Wicke S, Yi TS. 2019. Plastome reduction in the only parasitic gymnosperm Parasitaxus is due to losses of photosynthesis but not housekeeping genes and apparently involves the secondary gain of a large inverted repeat. Genome Biol Evol. 11(10):2789–2796.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Venkataramegowda S. 2012. Evaluation of Basella rubra L., Rumex nepalensis Spreng and Commelina benghalensis L. for antioxidant activity. Int J Pharm Pharm Sci. 4:714–720.
- Yi Y-L, Lei Y, Yin Y-B, Zhang H-Y, Wang G-X. 2012. The antialgal activity of 40 medicinal plants against Microcystis aeruginosa. J Appl Phycol. 24(4):847–856.