Abstract
The first complete chloroplast genome sequence of Sonchus arvensis, a herbaceous perennial, and its phylogenetic position relative to a member of the annual weedy and woody perennial species of Sonchus were reported in this study. Here, we assembled the complete plastome sequence of 151,967 base pairs (bp) in length, comprising 84,251 bp of a large single copy (LSC) and 18,184 bp (SSC) of small single copy confined between 24,766 bp of inverted repeats (IR). The genome contained 130 genes, including 87 protein-coding genes, six ribosomal RNA, and 37 transfer RNA genes. The overall GC content was 37.6% (LSC, 35.8%; SSC, 31.5%; IRs, 43.0%). Phylogenetic analysis confirmed that perennial S. arvensis is sister to the clade containing the weedy species of Sonchus.
Sonchus arvensis (milk field thistle, field or perennial sowthistle) belongs to the subgenus Sonchus, section Arvenses, and represents one of multiple recently radiated and highly unresolved lineages within the genus Sonchus (Kim et al. Citation2004, Citation2007; Mejías et al. Citation2018). Two infraspecific taxa, subsp. uliginosus (2n = 4x = 36) and subsp. arvensis (2n = 6x = 54), are currently recognized with native distributions in Europe and western Asia (Boulos Citation1973). Naturally occurring hybrids produced by the two subspecies have been detected in recently introduced areas where they are sympatric (Lemna and Messersmith 1990). As a noxious weed, S. arvensis has been rapidly introduced into various continents and countries, including North America, South America, Australia, Indonesia, Philippines, etc. In addition to being an early successional inhabitant on recently disturbed sites, S. arvensis is known for containing several active pharmaceutical compounds (Lemna and Messersmith Citation1990; Wahyuni et al. Citation2020). In the case of the phylogenetic position of S. arvensis, it is closely related to diploid (2n = 2x = 18) perennial species of Sonchus, including S. maritimus and S. crassifolius in sect. Maritimi and S. brachyotus in the same section Arvenses (Kim et al. Citation2007; Mejías et al. Citation2018). With regard to the plastome organization and evolution within the genus Sonchus, we have been generating the complete plastome sequences of woody perennials in the Canary Islands (Cho et al. Citation2019a), annual weedy species (Cho et al. Citation2019b), and one paleoendemic perennial species (Kim et al. Citation2019). Thus, we still know very little about the plastomes of other perennial species of Sonchus. As an effort to building a global plastome phylogenomic framework of Sonchus, we sequenced the complete plastome sequences of S. arvensis and assessed its phylogenetic position.
Total DNA (Voucher specimen: 41°14′27.9″N 83°41′51.1″W, OS418412) was isolated using the DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA) and sequenced by the Illumina HiSeq platform at Macrogen Corporation (Seoul, Korea). A total of 54,961,730 paired-end reads were assembled de novo with Velvet v. 1.2.10 using multiple k-mers (Zerbino and Birney Citation2008) and annotated by the Dual Organellar GenoMe Annotator (Wyman et al. Citation2004), ARAGORN v1.2.36 (Laslett and Canback Citation2004), and RNAmmer 1.2 Server (Lagesen et al. Citation2007).
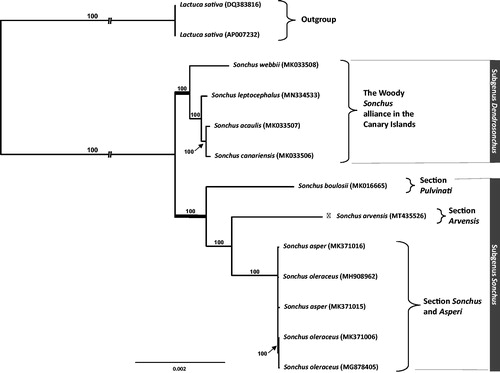
The complete chloroplast genome (GenBank: MT435526) of S. arvensis is 151,967 base pairs (bp) in length, containing a large single copy (LSC; 84,251 bp), a small single copy (SSC; 18,184 bp), and two inverted repeats (IRa and IRb; 24,766 bp each). The overall GC content was 37.6% (LSC, 35.8%; SSC, 31.5%; IRs, 43.0%) and the chloroplast contained 130 genes of which 87 were protein-coding genes, six coded for rRNA, and 37 for tRNA genes. To further investigate its phylogenetic position, eight representative species of Sonchus were aligned using MAFFT v.7 (Katoh and Standley, Citation2013) and maximum likelihood (ML) analysis was conducted using IQ-TREE v.1.4.2 (Nguyen et al. Citation2015). The ML tree showed that S. arvensis is sister to a clade containing members of the weedy Sonchus species, i.e., S. oleraceus and S. asper ().
Acknowledgments
We are grateful to UAB and SKKU for arranging and facilitating the exchange program and the degree of Genetics for including an External Practicum program. The first author thanks the Kim laboratory people at SKKU for their help during the course of this study.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank, National Center for for Biotechnology Information at (https://www.ncbi.nlm.nih.gov/genbank/), with reference number of MT435526.
Additional information
Funding
References
- Boulos L. 1973. Révision systématique du genre Sonchus L. s.l. IV. Sous-genre 1. Sonchus Bot Notiser. 126:155–196.
- Cho M-S, Kim JH, Kim C-S, Mejías JA, Kim S-C. 2019b. Sow thistle chloroplast genomes: insights into the plastome evolution and relationship of two weedy species, Sonchus asper and Sonchus oleraceus (Asteraceae). Genes. 10:881.
- Cho M-S, Yang JY, Yang T-J, Kim S-C. 2019a. Evolutionary comparison of the chloroplast genome in the woody Sonchus alliance (Asteraceae) on the Canary Islands. Genes. 10(3):217.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim S-C, Lee C, Mejías JA. 2007. Phylogenetic analysis of chloroplast DNA matK gene and ITS of nrDNA sequences reveals polyphyly of the genus Sonchus and new relationships among the subtribe Sonchinae (Asteraceae: Cichorieae). Mol Phylogenet Evol. 44(2):578–597.
- Kim S-C, Lu CT, Lepschi BJ. 2004. Phylogenetic positions of Actites megalocarpa and Sonchus hydrophilus (Sonchinae: Asteraceae) based on ITS and chloroplast non-coding DNA sequences. Aust Systematic Bot. 17(1):73–81.
- Kim S-H, Mejías JA, Kim S-C. 2019. Next generation sequencing reveals the complete plastome sequence of newly discovered cliff-dwelling Sonchus boulosii (Asteraceae: Cichorieae) in Morocco. Mitochondrial DNA B Resour. 4(1):164–165.
- Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNammer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35(9):3100–3108.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Lemna WK, Messersmith CG. 1990. The biology of Canadian weeds. 94. Sonchus arvensis L. Can J Plant Sci. 70(2):509–532.
- Mejías JA, Chambouleyron M, Kim S-H, Infante MD, Kim S-C, Léger J-F. 2018. Phylogenetic and morphological analysis of a new cliff-dwelling species reveals a remnant ancestral diversity and evolutionary parallelism in Sonchus (Asteraceae). Plant Syst Evol. 304(8):1023–1040.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Wahyuni DK, Purnobasuki H, Kuncoro EP, Ekasari W. 2020. Callus induction of Sonchus arvensis L. and its antiplasmodial activity. Afr J Infect Dis. 14(1):1–7.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.