Abstract
Renanthera imschootiana Rolfe is an epiphyte of genus Renanthera in family Orchidaceae. R. imschootiana is a rare tropical orchid with high ornamental value. We determined its complete chloroplast genome sequence to better understand its relationship with other Orchidaceae species. The chloroplast genome of R. imschootiana has a length of 143,836 bp, which comprises a large single copy region of 83,356 bp, a small single copy region of 11,172 bp, and a pair of reverse repeat regions of 24,654 bp each. Its genome has a total GC content of 36.8% and contains 127 genes, including 82 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Maximum likelihood phylogenetic analysis showed that R. imschootiana is closely related to Vanda brunnea, Holcoglossum flavescens, and H. amesianum.
Renanthera imschootiana has a high horticultural value and is widely distributed in Yuanjiang County, Yunnan Province, China. However, this species is endangered because of the over-exploitation of wild resources and the severe destruction of its original habitat. R. imschootiana has been listed as a critically endangered species in The China Biodiversity Red List: Higher Plants (Yang Citation2013) and appendix I species in the Convention on International Trade in Endangered Species of Wild Fauna and Flora (Li et al. Citation2018). Rebuilding a phylogenetic tree through high-throughput sequencing is necessary to understand the relationship between R. imschootiana and other Orchidaceae species.
The experimental materials of this study were collected from the wild plants of R. imschootiana in Xilahe Village, Yuanjiang County, Yunnan Province (101° 56′ 14″ E, 23° 44′ 36″ N, 1100 m). Test-tube seedlings were grown from their seeds by tissue culture, and DNA was extracted from the fresh leaves of the seedlings by modified CTAB method (Guan et al. Citation2004). The certificate was deposited at the Key Laboratory of Forest Resources Conservation and Utilization in the Southwest Mountains of China Ministry of Education, Southwest Forestry University (Accession Number: SWFU-SY36765). The complete chloroplast genome of R. imschootiana was assembled through the GetOrganelle software (Jin et al. Citation2018) using the publicly available Holcoglossum amesianum plasmid genome (Accession Number: NC_041511) as reference (Li et al. Citation2019) and was annotated in Geneious 8.1.3.
Results showed that the chloroplast genome of R. imschootiana is a double-linked DNA molecule with a length of 143,836 bp (LAU10049) and comprises a pair of 24,654 bp reverse repeat (IR) regions, a 83,356 bp large single copy (LSC) region, and a 11,172 bp small single copy (SSC) region. Its total GC content is 36.8% (LSC, 34.1%; SSC, 28.0%; IR, 43.4%). Its genome is larger by 3,027 and 4,238 bp than those of Holcoglossum flavescens (146,863 bp, NC_041512) and H. amesianum (148,074 bp, NC_041511), respectively. The chloroplast genome of R. imschootiana encodes 127 genes, namely, 82 protein-coding genes, 8 rRNA genes, and 37 tRNA genes.
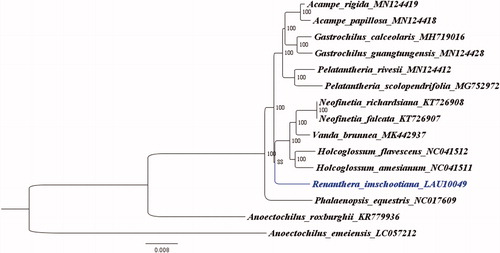
We reconstructed a phylogenetic tree () based on the published chloroplast genome sequences of 14 Orchidaceae species to confirm the evolutionary relationship between R. imschootiana and other Orchidaceae species . The chloroplast genome sequence of Anoectochilus emeiensis (Accession Number: LC057212) was used as an out-group, and the genome sequences were aligned using the MAFFT program version 7 (Katoh and Standley Citation2013). A maximum likelihood (ML) analysis based on the TVM + F+R2 model was performed through the IQ-TREE software version 1.6.7 using 1000 bootstrap replicates (Nguyen et al. Citation2015). The ML phylogenetic tree with 88–100% bootstrap values at each node indicated that R. imschootiana is closely related to Vanda brunnea, H. flavescens, and H. amesianum.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The chloroplast data of the R. imschootiana will be submitted to Lauraceae Chloroplast Genome Database (https://lcgdb.wordpress.com). Accession numbers are LAU10049.
Additional information
Funding
References
- Guan P, Kang JC, Zhou R, Shi JM. 2004. Comparative analysis of genomic DNA extraction methods in Gastrodia elata B1 in Orchidaceae. J Mount Agric Biol. 23(5):422–425.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assemble of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479. DOI: 10.1101/256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li J, Shi M, Lv YM, Yang JC, Du F. 2018. Discovery of an extremely endangered species Renanthera imschootiana and its population situation. Forest Res. 31(3):9–14.
- Li ZH, Ma X, Wang DY, Li YX, Wang CW, Jin XH. 2019. Evolution of plastid genomes of Holcoglossum (Orchidaceae) with recent radiation. BMC Evol Biol. 19(1):1–10.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Yang MS. 2013. The China Biodiversity Red List:Higher Plants. Beijing: China Environmental Yearbook; p. 646.