Abstract
Wrightia laevis Hook. f. is a great tree of Apocynaceae. It is mainly distributed in Southeast provinces of China and Southeast Asian countries. It is a plant that combines dyestuff and economic value. There is no study on the genome of W. laevisso far. Here we report and characterize the complete plastid genome sequence of W. laevis in order to provide genomic resources useful for promoting its conservation. The complete chloroplast genome of W. laevis is 155,274 bp in length with a typical quadripartite structure, consisting of a large single-copy region (LSC, 85,463 bp), a single-copy region (SSC, 18,181 bp) and a pair of inverted repeats (IRs, 25,815 bp). There are 133 genes annotated, including 88 unique protein-coding genes, 8 unique ribosomal RNA genes, and 37 transfer RNA genes. The overall G/C content in the plastome of W. laevis is 38.05%. The complete plastome sequence of W. laevis will provide a useful resource for the conservation genetics of this species as well as for phylogenetic studies in Apocynaceae.
Wrightia laevis Hook. f. is a plant of the family Apocynaceae, mainly distributed in Southeast provinces of China and Southeast Asian countries. It is a plant that combines medicinal and dyestuff value (Li et al. Citation1995). The chloroplast genome sequence carries rich information for plant molecular systematics and Barcoding. There have been no studies on the genome of W. laevis up to now. To provide a rich genetic information and improve W. laevis molecular breeding in the future, we report and characterize the complete plastid genome sequence of W. Laevis (GenBank accession number: MT505711).
In this study, the fresh leaves of W. laevis were collected from Jianfeng mountain in Hainan province (108.89° E, 18.73° N). A voucher specimens (HUTB 187223) were deposited in the Herbarium of the Institute of Tropical Agriculture and Forestry (code of herbarium: HUTB), Hainan University, Haikou, China.
The experiment procedure was as reported in Wang et al. (Citation2019). Total DNA of the W. laevis was sequenced with second-generation sequencing technology (Illumina HiSeq 2000, San Diego, CA). The chloroplast genome sequence reads were assembled with bioinformatic pipeline including SOAP2 software (Li et al. Citation2009) and several runs of manual corrections of sequence reads. Genes encoded by this genome were annotated by import the fasta format sequence to the DOGMA (Wyman et al. Citation2004) and recorrected by manual. The results showed that plastome of W. laevis possess a total length 155,274 bp with the typical quadripartite structure of angiosperms, containing two inverted repeats (IRs) of 25,815 bp, a large single copy (LSC) region of 85,463 bp and a small single copy (SSC) region of 18,181 bp. The plastome contains 133 genes, consisting of 88 unique protein-coding genes, 37 unique tRNA genes and 8 unique rRNA genes. The overall G/C content in the plastome of W. laevis is 38.05%, which the corresponding value of the LSC, SSC, and IR region were 36.20, 31.80, and 43.40%, respectively.
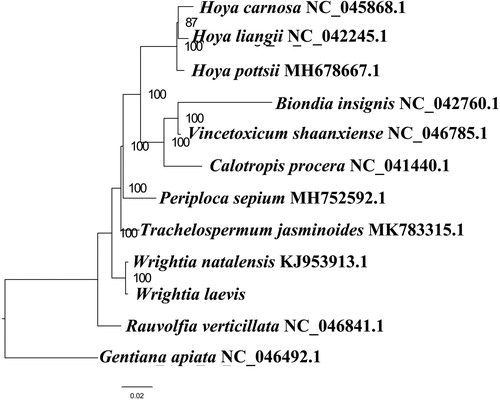
We used RAxML (Stamatakis Citation2006) with 1000 bootstraps under the GTRGAMMAI substitution model to reconstruct a maximum likelihood (ML) phylogeny of 12 published complete plastomes of Apocynaceae, using Gentianaapiata (Gentianaceae) as outgroups. According to the phylogenetic topologies, W. laevis was closely related to W. natalensis. Most nodes in the plastome ML trees were strongly supported (). The complete plastome sequence of W. laevis will provides a useful resource for the conservation genetics of this species as well as for the phylogenetic studies for Apocynaceae.
Figure 1. Maximum likelihood phylogenetic tree based on 12 complete chloroplast genomes. Accession number: Wrightia laevis (this study); Biondia insignis NC_042760.1; Calotropis procera NC_041440.1; Hoya carnosa NC_045868.1; Hoya liangii NC_042245.1; Hoya pottsii MH678667.1; Periploca sepium MH752592.1; Rauvolfia verticillata NC_046841.1; Trachelospermum jasminoides MK783315.1; Vincetoxicum shaanxiense NC_046785.1; Wrightia natalensis KJ953913.1; outgroup: Gentiana apiata NC_046492.1.The number on each node indicates the bootstrap value.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at http://www.ncbi.nlm.nih.gov, reference number MT505711.
Additional information
Funding
References
- Li PT, Antony JML, David JM. 1995. Flora of China. Beijing, People’s Republic of China: Science Press.
- Li RQ, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 25(15):1966–1967.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Wang HX, Chen L, Cheng XL, Chen WS, Li LM. 2019. Complete plastome sequence of Callicarpa nudiflora Vahl (Verbenaceae): a medicinal plant. Mitochondrial DNA B. 4(2):2090–2091.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
