Abstract
Elaeagnus is a genus which consists about 70 species of flowering plants in the family Elaeagnaceae, and its edible fruit is a natural product used as food and in traditional medicine. In this study, we sequenced the complete chloroplast (cp) genome of four species, namely Elaeagnus umbellate Thunb., E. multiflora Thunb., E. macrophylla Thunb., and E. glabra Thunb., to study their phylogenetic relationships within the Elaeagnaceae. Total lengths of the chloroplast genome were 152,261 bp, 152,267 bp, 152,224 bp, and 152,227 bp, respectively. The four genomes had representative quadripartite structures, with an LSC region (82,207 bp, 82,191 bp, 82,136 bp, and 82,139 bp) and an SSC region (18,262 bp, 18,282 bp,and 18,278 bp for both species) separated by a pair of IRs (25,896 bp, 25,897 bp, and 25,905 bp for the latter two species), respectively. Moreover, they were composed of 136–137 genes, including 88 protein-coding genes, 40–41 tRNA genes, and 8 rRNA genes. A maximum likelihood phylogenetic analysis indicated that E. umbellata was most closely related to E. multiflora, whereas E. macrophylla was close to E. glabra.
The genus Elaeagnus of the family Elaeagnaceae are plants that are distributed worldwide but are native mainly to temperate and subtropical regions of Asia, North America, southeastern Europe and Australia (Patel Citation2015). Many Elaeagnus species are used in health food and folk medicine. Among these, E. umbellata, E. multiflora, E. macrophylla, and E. glabraare are widely distributed in Korea and commonly consumed as food and herbal medicine (Shin et al. Citation2008; Li et al. Citation2009; Kim et al. Citation2016). Most research on Elaeagnus has focused on the aspects of commercial and industrial applications, such as their chemical and nutritional compositions. Studies on the molecular genetic diversity in the Elaeagnus genus are scarce. In this study, we used the Illumina Hiseq X platform to sequence the complete chloroplast (cp) genomes of four species: E. umbellata, E. multiflora, E. macrophylla, and E. glabra.
Fresh leaf material from four species were collected from Naju-si (35°00′22.0ʺN 126°49′27.8ʺE) in Jeollanam-do (Province), Korea. The specimens were deposited at the herbarium of the Jeollanam-do Forest Resources Institute (JFRI), Korea (specimen code JFRI00342741, JFR00342742, JFR00342743, and JFR00342744, respectively).
Total genomic DNA wa sextracted from fresh leaves by the CTAB method, and SEEDERS Inc. (Daejeon, Korea) performed whole-genome resequencing with the Illumina HiseqX Ten platform. Raw reads were filtered using Dynamic Trim and Length Short of the SolexaQA (ver.1.13) package (Cox et al. Citation2010).
Clean reads were first aligned to the cp genome of E. macrophylla (GenBank Accession No. NC_028066). Filtered reads were assembled into contigs in the software NOVOPlastyv 2.7.1 (Dierckxsens et al. Citation2017) and Fast-Plast v1.2.8 (Mckain et al. Citation2018). The physical maps of the new chloroplast genomes were generated using OGDRAWv 1.3.1 (Greiner et al. Citation2019). Finally, the validated complete cp genome sequences were submitted to the GenBank with accession numbers LC522506, LC523635, LC522136, and LC522137, respectively.
Total lengths of the chloroplast genome sequences of E. umbellata, E. multiflora, E. macrophylla, and E. glabra were 152,261 bp, 152,267 bp, 152,224 bp, and 152,227 bp, respectively. For E. umbellata, a pair of inverted repeats (IRs) of 25,896 bp was separated by a small single-copy (SSC) region of 18,262 bp and a large single-copy (LSC) region of 82,207 bp. The E. multiflora cp genome consisted of a LSC region of 82,191 bp, a SSC region of 18,282 bp, and IRs of 25,897 bp. Elaeagnus Macrophylla contained a LSC region of 82,136 bp, a SSC region of 18,278 bp, and IRs of 25,905 bp.The E. glabra cp genome consisted of a LSC region of 82,139 bp, a SSC region of 18,278 bp, and IRs of 25,905 bp. These new sequences each had a total of 137 genes (except for E. umbellata with 136 genes), including 88 protein-coding genes, 41 tRNA genes (except for E. umbellate with 40 genes), and 8 rRNA genes. The overall GC content of each whole plastome was 37.1%.
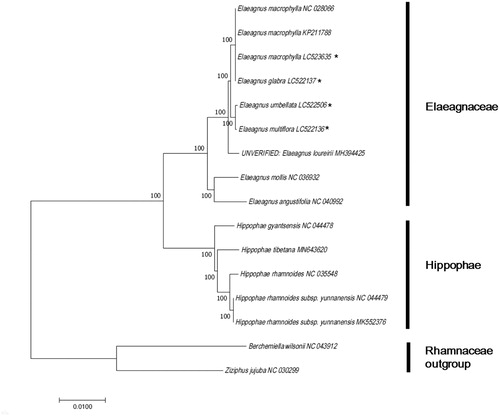
To investigate the phylogeny of E. umbellata, E. multiflora, E. macrophylla, and E. glabra, nine complete cp genomes belonging to Elaeagnaceae and related families were downloaded from GenBank and aligned using ClustalW (Thompson et al. Citation1994). A maximum likelihood method tree based on the Tamura–Nei model was constructed using MEGA 7.0 (Kumar et al. Citation2016). bThe ML tree analysis showed that E. umbellata had a closer relationship with E. multiflora; meanwhile, E. macrophylla was more closely related to E. glabra, with 100% bootstrap support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study openly available in National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession numbers LC522506, LC523635, LC522136, and LC522137, respectively.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 11(1):485.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e18.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Kim MJ, Lim JS, Yang SA. 2016. Component analysis and anti-proliferative effects of ethanol extracts of fruits, leaves, and stems from Elaeagnus umbellata in HepG2 cells. J Korean Soc Food Sci Nutr. 45(6):828–834.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li L-H, Baek IK, Kim JH, Kang KH, Koh YS, Jung YD, Cho CK, Choi S-Y, Shin BA. 2009. Methanol extract of Elaeagnus glabra, a Korean medicinal plant, inhibits HT1080 tumor cell invasion. Oncol Rep. 21(2):559–563.
- McKain MR, Wilson MC, Kellogg EA. 2018. Fast-Plast: a rapid de novo assembly pipeline for whole chloroplast genomes. Git-Hub. 973887. Doi: 10.5281/zenodo
- Patel S. 2015. Plant genus Elaeagnus: underutilized lycopene and linoleic acid reserve with permaculture potential. Fruits. 70(4):191–199.
- Shin SR, Hong JY, Yoon KY. 2008. Antioxidant properties and total phenolic contents of cherry elaeagnus (Elaeagnus multifloraThunb.) leaf extracts. Food Sci Biotechnol. 17(3):608–612.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680.