Abstract
Ficus altissima Blume is a hemiepiphytic monoecious fig species of the genus Ficus in the family Moraceae. To better determine its phylogenetic location with respect to the other Ficus species, the complete plastid genome of F. altissima was sequenced. The whole plastome is 160,251 bp in length, consisting of a pair of inverted repeat (IR) regions of 25,886 bp, one large single-copy (LSC) region of 88,470 bp, and one small single-copy (SSC) region of 20,009 bp. The overall GC content of the whole plastome is 35.9%. Further, maximum likelihood phylogenetic analyses was conducted using 29 complete fig plastomes, which support close relationships among F. altissima, F. benjamina, F. microcarpa, and F. consociata.
Ficus altissima Blume, a hemiepiphytic species distributed in tropical and subtropical areas of much of Asia, was assigned to subgenus Urostigma section Conosycea in the family Moraceae (Berg and Corner Citation2005). The figs of F. altissima are occupied by two Eupristina species in southern China: the pollinator Eupristina altissima and the cheater Eupristina sp. (Zhao et al. Citation2014; Chen et al. Citation2018). Adult females of both species share similar biological characteristics but showed different reproductive strategies (Xu et al. Citation2016). For a better understanding of the relationships of F. altissima and other fig species, it is necessary to reconstruct a phylogenetic tree based on high-throughput sequencing approaches.
Fresh young leaves of F. altissima in Xishuangbanna (Yunnan, China; Long. 101.2611 E, Lat. 21.9275 N, 555 m) were picked for DNA extraction (Doyle and Dickson Citation1987). The voucher was deposited at the Biodiversity Research Group of Xishuangbanna Tropical Botanical Garden (Accession Number: XTBG-BRG-10001). The whole plastid genome was sequenced following Zhang et al. (Citation2016), and their 15 universal primer pairs were used to perform long-range PCR for next-generation sequencing. The contigs were aligned with with the cp genomes of relatively related species and annotated in Geneious 4.8.
The plastome of F. altissima (LAU10101), with a length of 160,251 bp, was 778 bp larger than that of F. racemosa (159,473 bp, KT368151). It was also 123 bp, 351 bp and 376 bp smaller than that of F. hirta (160,374 bp, MN364706), F. carica (160,602 bp, KY635880) and F. religiosa (160,627 bp, KY416513). The length of the inverted repeats (IRs), large single-copy (LSC), and small single-copy (SSC) regions of F. altissima was 25,886 bp, 88,470 bp, and 20,009 bp, respectively. The overall G + C content is 35.9% (LSC, 33.6%; SSC, 28.8%; IR, 42.6%).
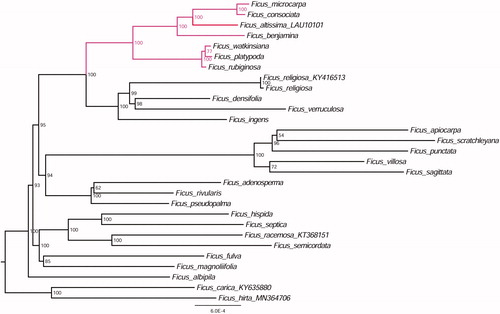
Furthermore, based on 29 published plastid genome sequences (Bruun-Lund et al. Citation2017), we reconstructed a phylogenetic tree () to confirm the evolutionary relationship between F. altissima and other species with published plastomes in Ficus. Maximum likelihood (ML) phylogenetic analyses were performed based on GTR + F + R3 model in the iqtree version 1.6.7 program with 1000 bootstrap replicates (Nguyen et al. Citation2015). The ML phylogenetic tree with 54–100% bootstrap values at each node supported that F. altissima was closely related to and F. benjamina, F. consociate, and F. microcarpa.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the finding of this study are openly available in Moraceae Chloroplast Genome Database (http://lcgdb.wordpress.com). Accession numbers are LAU10101.
Additional information
Funding
References
- Berg CC, Corner E. 2005. Moraceae (Ficus). In: Nooteboom HD, editor. Flora Malesiana, Series I, Vol 17(2). Leiden, The Netherlands: National Herbarium of Nederland; p. 1–625.
- Bruun-Lund S, Clement WL, Kjellberg F, Rønsted N. 2017. First plastid phylogenomic study reveals potential cyto-nuclear discordance in the evolutionary history of Ficus L. (Moraceae). Mol Phylogenet Evol. 109:93–104.
- Chen HH, Zhang Y, Peng YQ, Corlett RT. 2018. Latitudinal effects on phenology near the northern limit of figs in china. Sci Rep. 8(1):4320.
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36(4):715–722.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Xu R, Zhang Y, Peng YQ, Yang DR. 2016. Reproductive characteristics of pollinator and cheater wasps that utilize the female flowers of Ficus altissima. Acta Ecol Sin. 36:1134–1140.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Sytemat Evol. 54(3):219–227.
- Zhao JB, Peng YQ, Quinnell RJ, Compton SG, Yang DR. 2014. A switch from mutualist to exploiter is reflected in smaller egg loads and increased larval mortalities in a ‘cheater’ fig wasp. Acta Oecol. 57:51–57.