Abstract
Here, we present the complete mitochondrial genome of Lophosquillia costata. The genome is 15,771 bp in length with a 68.07% AT content. It contains 13 protein-coding genes, two rRNAs genes, and 22 tRNAs. Both rRNAs are encoded on the light strand. Besides seven tRNAs are encoded on the light strand (trnY, trnQ, trnV, trnL1, trnP, trnH, and trnF), and four PCG (nad1, nad4l, nad4, and nad5) are encoded on the light strand, whereas the other nine PCGs are located on the heavy strand. Phylogenetic analysis based on mitochondrial PCGs shows two distinct groups for Stomatopoda and Decapoda. Lophosquillia costata is found clustered with Oratosquilla oratoria into a branch (BP = 100), and they grouped with other species with high support (BP = 99) in the family Squillidae. Our results shall provide a better understanding in the evolutionary histories of the stomatopods.
Stomatopods form an important component of marine ecosystems and are economically significant (Van Der Wal et al. Citation2019). The largest superfamily the order of Stomatopoda is Squilloidea, which is morphologically diverse and includes over 185 species in 49 genera (Ahyong Citation2001, Citation2005; Van Der Wal and Ahyong Citation2017). As active predators in muddy and sandy substrates on coastal and continental shelf habitats (Abelló and Martin Citation1993; Ahyong Citation2005), some species of Squilloidea are also major fisheries targets, including Squilla mantis (Abelló and Martin Citation1993; Maynou et al. Citation2004), Oratosquilla oratoria (Zhang et al. Citation2012) and Harpiosquilla raphidea (Wardiatno and Mashar Citation2010). Studies on Squilloidea have been focused on Oratosquilla oratoria in China (Zhang et al. Citation2012, Citation2016; Yang and Li Citation2018), and here we report on another species, Lophosquilla costata (de Haan, 1844) (Crustacea: Malacostraca: Stomatopoda: Squillidae), one of the dominant species of mantis shrimps in the Zhoushan Fishing Ground and its adjacent waters (Yu et al. Citation2011).
As of 15 March 2020, GenBank contained seven complete mitochondrial genomes of Stomatopoda, including three families. There is one partial mitochondrial sequence (GenBank: MH168237.1) of Lophosquilla costata in Genbank, however, the whole mitochondrial genome is still in lack. Here, we present the first complete mitochondrial genome of the L. costata.
Specimens of L. costata were collected from Dongfushan Island (30.13°N, 122.77°E), Zhoushan archipelago, in the East China Sea. The muscle tissue isolated from the fresh specimen was immediately preserved in 95% ethanol and kept in −80 °C. DNA was extracted with E.Z.N.A®DNA kit (OMEGA, USA), and mitochondrial DNA was amplified with a DNA REPLI-g Mitochondrial DNA Kit (QIAGEN, Hilden, Germany) as directed by the manufacturer. Library construction and sequencing were performed by Biozeron (Biozeron, Shanghai, China) using the Illumina HiSeq 4000 sequencing platform (Illumina, San Diego, CA). The specimen is stored in −80 °C in Fishery resources lab in Marine and Fisheries Research Institute of Zhejiang Province (Mshrimp MT-1).
The mitochondrial genome of L. costata is a circular molecule which is 15,771 bp (GenBank accession number: MT276143) in length. It contains 13 protein-coding genes (PCGs), two rRNAs genes, and 22 tRNAs. Total AT content of L. costata is 68.07%. Both rRNAs are encoded on the light strand. Besides seven tRNAs are encoded on the light strand (trnY, trnQ, trnV, trnL1, trnP, trnH, and trnF), and four PCG (nad1, nad4l, nad4, and nad5) are encoded on the light strand, whereas the other nine PCGs are located on the heavy strand.
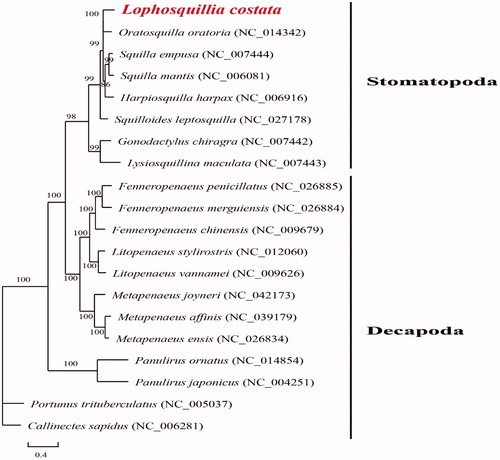
To elucidate phylogenetic relationships of L. costata with the other stomatopods, phylogenetic tree () is constructed based on the PCGs with Maximum-Likelihood using phyML ver 3.0 (http://www.atgc-montpellier.fr/phyml/). The genetic information from Order Stomatopoda has only been reported several times (e.g. Kundu et al. Citation2018), those from Order Decapoda are also considered (e.g. Sung et al. Citation2018). A total of six mitochondrial genomes from Stomatopoda and 12 mitochondrial genomes from Decapoda have been used in the phylogenetic tree. All the six species from Order Stomatopoda were belonged to the same family except Lysiosquillina maculata (Lysiosquillidae) and Gonodactylus chiragra (Gonodactylidae). Results show two distinct groups for Stomatopoda and Decapoda. Lophosquillia costata is found clustered with Oratosquilla oratoria into a branch (BP = 100), and they grouped with other species with high support (BP = 99) in the family Squillidae. Two species from other families, Lysiosquillina maculata and Gonodactylus chiragra, are the most distantly related species within Stomatopoda, also with high support (BP = 99).
Figure 1. Phylogenetic tree of Lophosquillia costata and other genomes from Order Stomatopoda and Decapoda based on mitochondrial PCGs.
In this study, we present the complete mitochondrial genome sequence of L. costata, which would contribute to further phylogenetic analysis of this species. Furthermore, more mitochondrial genomic data of undetermined taxa and further analysis are required to reveal phylogeny and evolution of stomatopods.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in the Genbank database at https://www.ncbi.nlm.nih.gov/ (Accession number: MT276143) after 31 May 2021, and also available from the corresponding author [YX. Bi], upon reasonable request.
Additional information
Funding
References
- Abelló P, Martin P. 1993. Fishery dynamics of the mantis shrimp Squilla mantis (Crustacea: Stomatopoda) population off the Ebro delta (northwestern Mediterranean). Fish Res. 16(2):131–145.
- Ahyong ST. 2001. Revision of the Australian stomatopod Crustacea. Rec Aust Mus, Suppl. 26(Supplement):1–310.
- Ahyong ST. 2005. Phylogenetic analysis of the Squilloidea (Crustacea: Stomatopoda). Invert Syst. 19(3):189–208.
- Kundu S, Rath S, Tyagi K, Chakraborty R, Pakrashi A, Kumar V, Chandra K. 2018. DNA barcoding of Cloridopsis immaculata: genetic distance and phylogeny of stomatopods. Mitochondrial DNA B. 3(2):955–958.
- Maynou F, Abello P, Sartor P. 2004. A review of the fisheries biology of the mantis shrimp, Squilla mantis (L., 1758) (Stomatopoda, Squillidae) in the Mediterranean. Crustac. 77(9):1081–1099.
- Sung CH, Cheng CC, Lu JK, Wang LJ. 2018. The complete mitochondrial genome of Hymenocera picta (Malacostraca: Decapoda: Hymenoceridae). Mitochondrial DNA B. 3(2):790–791.
- Van Der Wal C, Ahyong ST, Ho SYW, Lins LSF, Lo N. 2019. Combining morphological and molecular data resolves the phylogeny of Squilloidea (Crustacea: Malacostraca). Invert Syst. 33:89–100.
- Van Der Wal C, Ahyong ST. 2017. Expanding diversity in the mantis shrimps: two new genera from the eastern and western Pacific (Crustacea:Stomatopoda:Squillidae). Nauplius. 25(0):e2017012.
- Wardiatno Y, Mashar A. 2010. Biological information on the mantis shrimp, Harpiosquilla raphidea (Fabricius 1798) (Stomatopoda, Crustacea) in Indonesia with a highlight of its reproductive aspects. Journal of Tropical Biology & Conservation. 7:65–73.
- Yang M, Li XZ. 2018. Population genetic structure of the mantis shrimp Oratosquilla oratoria (Crustacea: Squillidae) in the Yellow Sea and East China Sea. J Ocean Limnol. 36(3):905–912.
- Yu CG, Chen QZ, Chen XQ, Ning P, Zheng J. 2011. Species composition and quantity profile of mantis shrimp around Zhoushan fishing ground. J Dalian Ocean Univ. 26(2):153–156. (in Chinese with English abstract).
- Zhang D, Ding G, Ge B, Liu Q, Zhang H, Tang B, Yang G. 2016. Geographical distribution, dispersal and genetic divergence of the mantis shrimp Oratosquilla oratoria (Stomatopoda: Squillidae) in China Sea. Biochem Syst Ecol. 65:1–8.
- Zhang DZ, Ding G, Ge BM, Zhang HB, Tang BP. 2012. Development and characterization of microsatellite loci of Oratosquilla oratoria (Crustacea: Squillidae). Conservation Genet Resour. 4(1):147–150.
