Abstract
Apocynum venetum L. (Apocynaceae) or Luobuma is a widely known traditional medicine use to treat hypertension, relieve anxiety, soothe the nerves and promote diuresis. In this study, the complete chloroplast genome of this medicinal plant was determined through Illumina sequencing method. The A. venetum cp genome is 150,897 bp in length, containing a small single copy region (17,256 bp), a large single copy region (81,957 bp), and a pair of IR regions (25,842 bp). It encodes for a total of 131 genes, including 86 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. Phylogenetic analysis also reveals that A. venetum is relatively close to Aganosma cymosa.
Apocynum venetum L. (Apocynaceae), commonly known as Luobuma, is a traditional Uygur medicine popular in Xinjiang, China (Xie et al. Citation201 2). Various ethnopharmacological researches documented that A. venetum can successfully lower blood pressure and has antihepatotoxic, cardiotonic, anxiolytic and antioxidant activities (Xie et al. Citation2012). Luobuma tea has also been commercially recognized as an anti-hypertensive, anti-platelet aggregation, anti-depressant, anti-aging, and sedative nutritional supplement thus gaining its popularity in China, Japan and American health food markets (Xie et al. Citation2012; Gao et al. Citation2019). But despite its widely held medicinal value, genomic information about this plant is still lacking. Closely related herb Apocynum pictum Schrenk is morphologically similar and easily confused for A. venetum, which can possibly cause misconception and problems in medicine and health product markets (Xie et al. Citation2012). Hence, this report presents the complete chloroplast genome sequence of A. venetum to provide basic genetic resource that can be used in authenticating this highly medicinal plant species.
Genomic DNA was extracted using the CTAB method from five grams of Apocynum venetum fresh leaves collected in Keziertuoan, Jinghe County, Bortala Mongol Autonomous Prefecture, Xinjiang, China (N44.4440; E82.1680). Voucher specimen (Chen et al. 2019062401) was deposited in the herbarium of Sun Yat-sen (Zhongshan) University (SYS). The genome sequencing was performed in Illumina Hiseq 2500 platform producing 150 bp paired end reads. High-quality reads were assembled into contigs via the de novo assembler SPAdes 3.11.0, using a k-mer set of 93, 105, 117, 121 (Nurk et al. Citation2013). The A. venetum cp genome protein coding genes annotation was conducted using CpGAVAS (Liu et al. Citation2012) while tRNA genes were annotated in tRNAscan-SE v2.0 (Chan and Lowe Citation2019).
The complete chloroplast genome of A. venetum (GenBank accession number: MT313688) exhibits a quadripartite structure with a length of 150,897 bp, including a large single copy (LSC) region (81,957 bp), a small single copy (SSC) region (17,256 bp) and two inverted repeats (IR) regions (25,842 bp). Overall GC content is 38.3%. A total of 131 genes were determined, including 86 protein-coding genes, 8 rRNA genes, and 37 tRNA genes. This accounts for the 61% of the whole A. venetum cp genome. Similar to other angiosperm cp genomes, A. venetum cp genome contains genes with introns. The genes trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, ndhA have one intron each while clpP, ycf3 contain two introns. Trans-splicing event was also observed in rps12 gene.
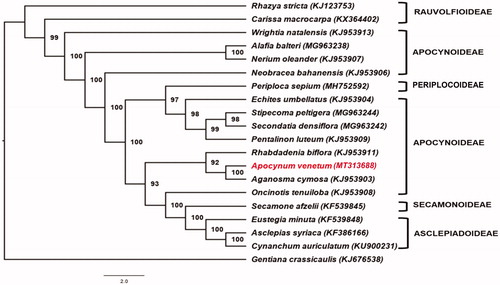
Phylogenetic analysis was carried out using RAxML 8.2.11 with the GTRI + G nucleotide substitution model (Stamatakis Citation2014). Protein coding regions of A. venetum cp genome and 19 other reported Apocynaceae cp genome sequences were used to reconstruct the phylogenetic tree. Analysis reveals that A. venetum is closely-related to Aganosma cymosa and both belong to tribe Apocyneae (). However, some relative positions did not support the traditional classification of Apocynaceae at the subfamily level. Most placements are consistent to the published phylogenetic tree presented by Rao et al. (Citation2019).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All the chloroplast genome sequences used in this research are available in GenBank database. The complete mitochondrial genome of Apocynum venetum is deposited in NCBI GenBank with the accession number MT313688. (https://www.ncbi.nlm.nih.gov/nuccore/MT313688)
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Gao G, Chen P, Chen J, Chen K, Wang X, Abubakar AS, Liu N, Yu C, Zhu A. 2019. Genomic survey, transcriptome, and metabolome analysis of Apocynum venetum and Apocynum hendersonii to reveal major flavonoid biosynthesis pathways. Metabolites. 9 (12):296.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, et al. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 20(10):714–737.
- Rao H, Song ZH, Cui RP, Li QE, Zou JB. 2019. Characterization of the complete chloroplast genome of Periploca sepium Bunge (Apocynaceae: Periplocoideae: Periploca), a traditional Chinese medicinal plant. Mitochondria DNA Part B. 4(1):335–336.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. 1
- Xie W, Zhang XY, Wang T, Hu J. 2012. Botany, traditional uses, phytochemistry and pharmacology of Apocynum venetum L. (Luobuma): a review. J Ethnopharmacol. 141(1):1–8.