ABSTACT
Cymbidium longibracteatum is a common cultivated species in the genus Cymbidium due to its elegant appearance, rich flower colors and strong fragrance, but its classification is quite controversial. In this study, the complete chloroplast genome of C. longibracteatum was obtained by Illumina sequencing. The chloroplast genome of C. longibracteatum is 150,070 bp in length with an overall GC content of 37.12%, which contains a large single-copy (LSC;84,949 bp) region, a small single-copy (SSC;13,745bp) region, and a pair of inverted repeats (IRs; 25,688 bp) regions. The genome contains 130 genes, namely 84 protein-coding genes, 38 tRNA genes and 8 rRNA genes. The maximum-likelihood phylogenetic tree has proved that C. longibracteatum should exist as an independent species in the genus Cymbidium, and it is most closely related to C. tortisepalum. This study provides valuable sequence resources for further study of C. longibracteatum.
Cymbidium longibracteatum (Orchidaceae) is mainly distributed in Sichuan, Guizhouand Yunnan Province of China, known as the authentic Sichuan orchid (Jie et al. Citation2013; Zhang et al. Citation2019). It exceeds most traditional and popular orchids owing to its elegant appearance, rich colors, and strong fragrance, which has high ornamental and economic value (Fengyan et al. Citation2009). Although C. longibracteatum is a common cultivated species in the genus Cymbidium, it is quite controversial in classification (Singchi and Zhongjian Citation2003). Yingsiang and Singchi (Citation1980) classified C. longibracteatum as a variety of C. goeringii. Singchi and Zhongjian (Citation2003), however, classified C. longibracteatum as a variety of C. tortisepalum. Due to the natural hybridization of the Cymbidium species in the nature, there are many intermediate types, making the boundaries of this species unclear (Ning et al. Citation2018). Today, the classification of Cymbidium species is mainly based on morphological indicators (Jiaping and Silan Citation1998). Meanwhile, there is also a lack of DNA data of the Cymbidium species, which is one of the most important tools in taxonomy (Ning et al. Citation2018). Therefore, it is urgent to provide valuable genetic information for this species. The complete chloroplast genome sequence can provide reliable data to identify species that are controversial in taxonomy, and it is shorter in length and more conservative in structure than the nuclear and mitochondrial genomes (Scarcelli et al. Citation2011). Here, we were the first to report the complete chloroplast genome of C. longibracteatum. This study will offer reliable molecular genetic data for the subsequent classification and identification of Cymbidium.
The mature leaves of C. longibracteatum were collected from Tang’jia mountain in Hongkou town, Bazhong city, Sichuan province, China (32°12′32.49″N; 107°58′26.89″E), and voucher specimen deposited at Orchid Resource Nursery of Zhejiang Agriculture and Forestry University (specimen code ZAFU20120218). Total genomic DNA was extracted by the modified CTAB method (Fu et al. Citation2017) and sequenced by NovaSeq platform (Illumina, USA). The clean reads were assembled by NOVOPlasty (Dierckxsens et al. Citation2017). The assembled sequence was annotated using CpGAVAS (Liu et al. Citation2012). The chloroplast genome map was generated using the online tool OGDRAW (Lohse et al. Citation2007). Finally, the complete chloroplast genome of C. longibracteatum was submitted to the GenBank (Accession Number: MT259022).
The chloroplast genome of C. longibracteatum is 150,070 bp in length with an overall GC content of 37.12%, which contains a large single-copy (LSC; 84,949 bp) region, a small single-copy (SSC; 13,745bp) region, and a pair of inverted repeats (IRs; 25,688 bp) regions. The genome encodes130 genes, namely 84 protein-coding genes, 38 tRNA genes, and 8rRNA genes.
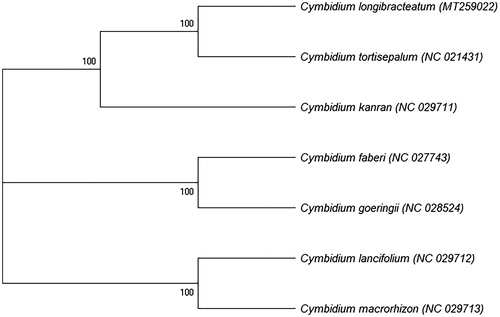
To determine the phylogenetic position of C. longibracteatum, we selected 7 complete chloroplast genomes sequence of Cymbidium from NCBI GenBank for phylogenetic analysis. The sequences were aligned using MEGA X (Kumar et al. Citation2018) and the maximum-likelihood (ML) tree was constructed using RAxML v8.2.12 (Stamatakis, Citation2014) with 1000 bootstraps (). The results indicated that C. longibracteatum should exist as an independent species in the genus Cymbidium, not as a variety of C. goeringii or C. tortisepalum, and it was sister to C. tortisepalum. The complete chloroplast genome of C. longibracteatum will contribute to further study of this species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov, GenBank Accession Number: MT259022.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18–e.
- Fengyan W, Chengxiu L, Changxian W, Dongxu Z, Yinping P. 2009. Study on propagation and differentiation of PLB of Cymbidium hybridum×Cymbidium Longibracteatum. Chin Agri Sci Bulletin. 25(23):327–330.
- Fu Z-y, Song J-c, Jameson PE. 2017. A rapid and cost effective protocol for plant genomic DNA isolation using regenerated silica columns in combination with CTAB extraction. J Integrat Agric. 16(8):1682–1688.
- Jiaping H, Silan D. 1998. The numerical taxonomy of Chinese cymbidium. J Beijing Forest Univ. 20(02):42–47.
- Jie L, Ping K, Na WZ, Zang YJ. 2013. Distribution status of wild Cymbidium tortisepalum var. longibracteatum resources and associated plants in parallel ridge-valley region in the eastern Sichuan of China. Resour Environ Yangtze Basin. 22(10):1319–1324.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Lohse M, Drechsel O, Bock R. 2007. Organellar GenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- Ning H, Ao S, Fan Y, Fu J, Xu C. 2018. Correlation analysis between the karyotypes and phenotypic traits of Chinese cymbidium cultivars. Hortic Environ Biotechnol. 59(1):93–103.
- Scarcelli N, Barnaud A, Eiserhardt W, Treier UA, Seveno M, d'Anfray A, Vigouroux Y, Pintaud J-C. 2011. A set of 100 chloroplast DNA primer pairs to study population genetics and phylogeny in monocotyledons. PLOS One. 6(5):e19954–e.
- Singchi C, Zhongjian L. 2003. Critical notes on some taxa of Cymbidium. Acta Phytotaxon Sin. 41(01):79–84.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yingsiang W, Singchi C. 1980. A taxonomic review of the orchid genus Cymbidium in China. Acta Phytotaxon Sin. 18(3):292–307.
- Zhang Q, Li J, Shan M, Liu X, Li Y. 2020. Microscopic observation on the process of axenic seed germination of Cymbidium tortisepalum var. longibracteatum. J Nanjing Forest Univer (Nat Sci Ed). 44(02):105-110.